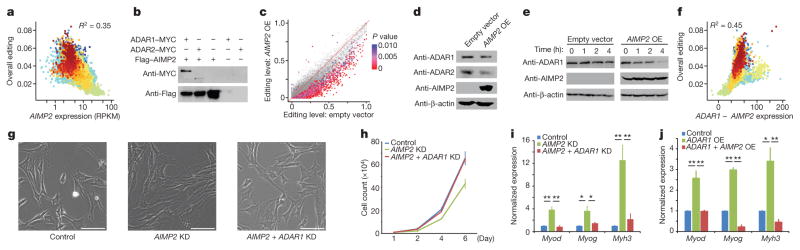
Figure 4. Identification of AIMP2 as a negative regulator of A-to-I editing.
a, Correlation of AIMP2 expression with overall editing of all sites in the GTEx samples. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. b, Co-immunoprecipitation experiment with either MYC-tagged ADAR1 or MYC-tagged ADAR2 and 3×Flag-tagged AIMP2 in HEK293T cells. Anti-Flag M2 beads were used to immunoprecipitate the regulator, and anti-MYC was then used to probe whether the relevant editing enzyme was pulled down together with AIMP2. c, Comparison of editing levels between control cells and cells with AIMP2 overexpressed (OE). The red-purple coloured dots indicate the differentially edited sites (P <0.01, Fisher’s exact test). d, Western blot analysis of ADAR1 and ADAR2 protein levels with or without overexpression of AIMP2 in HEK293T cells. Only the p110 isoform of ADAR1 was detected. e, Cycloheximide-chase analysis followed by western blotting was used to determine the rate at which the ADAR1 p110 protein was degraded with or without AIMP2 overexpression. f, Correlation of ADAR1 expression with overall editing of all sites in the GTEx samples when the negative influence of AIMP2 was taken into account. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. g–i, Effect of knocking down (KD) either AIMP2 alone or both AIMP2 and ADAR1 concurrently in undifferentiated C2C12 myoblasts. Morphology (g), proliferation rate (h) and expression (i) of muscle-specific markers. Myh3, myosin heavy chain; Myod, myogenic differentiation 1; Myog, myogenin. Scale bars, 100 μm. j, Effect of overexpressing ADAR1 alone or both ADAR1 and AIMP2 together in C2C12 myoblasts on the expression of muscle-specific markers. *P <0.05, **P <0.01, Student’s t-test. Error bars denote s.e.m. from three biological replicates.