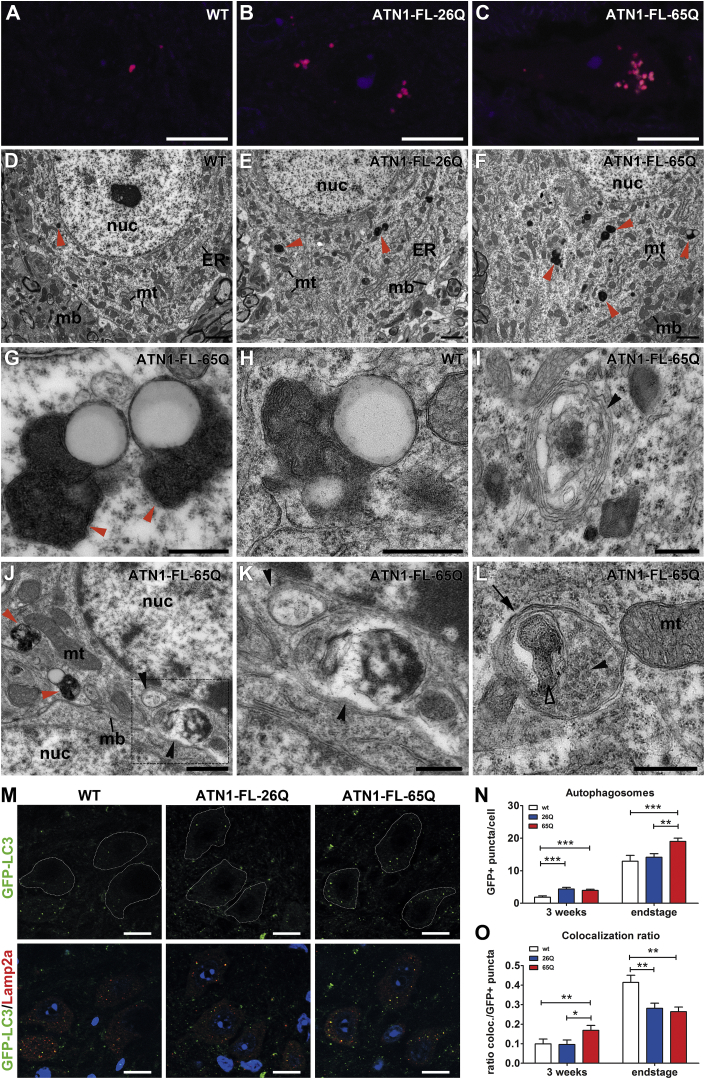
Figure 2.
Accumulation of Undigested Autophagic Structures in Cells of the Dentate Nucleus at End-Stage in DRPLA Mice
(A–C) Increased accumulation of lipofuscin-like autofluorescence in the dentate nucleus of ATN1-FL-65Q (C) and a lower level of increase in ATN1-FL-26Q (B) mice compared to wild-type (A). Confocal laser scanning microscope image λex = 514 and 633. Scale bar, 10 μm. For quantification, see Figure S1G; for examples of autofluorescence bleaching, see Figure S2A.
(D–F) Low-magnification TEM images of DN cells show an accumulation of tertiary lysosomes as electron dense vesicular structures with transparent lipid inclusions, known as lipofuscin (red arrowheads) in ATN1-FL-65Q mice (F) compared to wild-type (WT, D) and ATN1-FL-26Q (E). nuc, nucleus; mt, mitochondria; ER, endoplasmatic reticulum; mb, plasma membrane. Scale bar, 1 μm. Higher-magnification image of (F) is shown in Figure 5B.
(G and H) High-magnification TEM images of tertiary (late) lysosomes resemble the typical structure of lipofuscin-packed electron dense lamellar matrix containing intravesicular lipid inclusion as an electron transparent circular structure. (G) ATN1-FL-65Q and (H) wild-type (WT). mt, mitochondria. Scale bar, 500 nm.
(I) High-magnification TEM image shows dense accumulation surrounded by multiple membranes (black arrowhead) as observed in ATN1-FL-65Q dentate nucleus cells, often referred to as a multilamellar body. Scale bar, 500 nm
(J) Example of a cell showing multiple accumulations of tertiary lysosomes (red arrowheads) and double-membrane autophagic vesicles (black arrowheads) containing undigested debris frequently observed in ATN1-FL-65Q dentate nucleus cells. Scale bar, 1 μm. Lower-magnification representation of the nucleus is shown in Figure 5A.
(K) Magnification of the inset in (J) showing two double-membrane vesicular structures (black arrowheads), containing undigested material. Scale bar, 500 nm.
(L) High-magnification image of an autophagosome (arrow), as a double-membrane vesicular structure containing several endosomes (full arrowhead), and an undigested former mitochondrion (framed arrowhead). Scale bar, 500 nm.
(M) Representative images of dentate nucleus cells from WT;GFP-LC3, ATN1-FL-26Q;GFP-LC3, and ATN1-FL-65Q;GFP-LC3 end-stage mice evaluated for GFP (green) and LAMP2a (red) positive as well as colocalized (yellow) puncta. Puncta were counted within the soma of cells, showing typical morphology for DN neurons with large low-intensity nuclei (blue) surrounded by a relatively large cytoplasm and a high LAMP2a positive background. Images were taken at the nuclear level of the cell with Axiovert epifluorescence microscope using Apotome optical sectioning with high grid at 100× objective magnification. Scale bar, 10 μm.
(N) Quantification of GFP-positive puncta in dentate nucleus cells of WT;GFP-LC3 (wt), ATN1-FL-26Q;GFP-LC3 (26Q), and ATN1-FL-65Q;GFP-LC3 (65Q) mice at the presymptomatic stage of 3 weeks (wt, n = 83 cells, 3 animals; 26Q, n = 77 cells, 3 animals; 65Q, n = 96 cells, 4 animals) and end-stage (wt, n = 53 cells, 3 animals; 26Q, n = 49 cells, 3 animals; 65Q, n = 52 cells, 3 animals). One-way ANOVA, mean ± SEM, ∗∗∗p < 0.001, ∗∗p < 0.01.
(O) Relative co-localization ratio of puncta positive for both GFP-LC3 and LAMP2a to puncta positive for only GFP-LC3 in dentate nucleus cells of 3 week and end-stage WT;GFP-LC3 (wt), ATN1-FL-26Q;GFP-LC3 (26Q) and ATN1-FL-65Q;GFP-LC3 (65Q) mice. One-way ANOVA, mean ± SEM, ∗∗p < 0.01, ∗p < 0.05.
See also Figure S2.