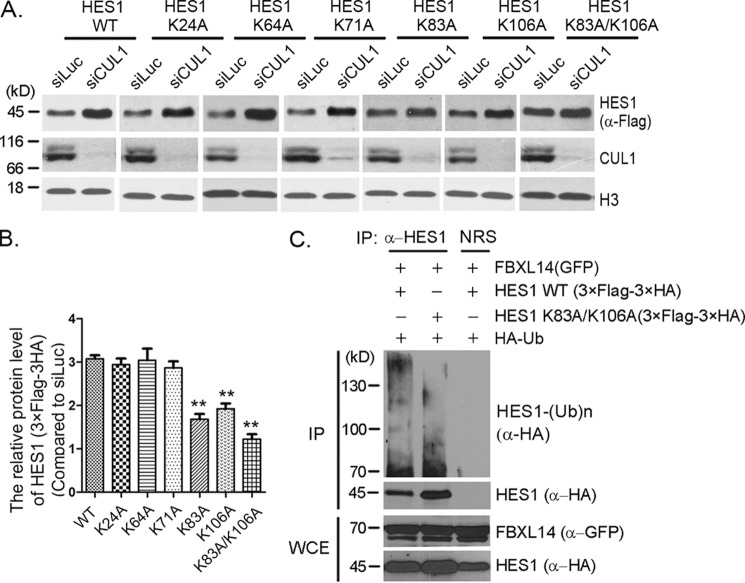
Figure 5.
Lysines 83 and 106 were potential ubiquitination sites for SCFFBXL14-mediated HES1 ubiquitination. A, lysine mutants screening. Stably expressing 3× FLAG–3× HA–HES1 WT or mutants (as indicated) F9 cells were transfected with luciferase or CUL1 siRNAs for 45 h, and cell lysates were immunoblotted using indicated antibodies. Histone H3 was taken as loading control. B, quantification of the relative protein levels of HES1 and mutants in A. The quantification of protein levels was measured as in Fig. 1A. The statistical difference between the control group (WT) and experimental groups (K24A, K64A, K71A, K83A, K106A, and K83A/K106A) was measured. The error bars represent S.D. from three independent experiments. The significance of statistical difference was calculated by paired two-sided Student's t test (**, p < 0.01). C, HES1 was ubiquitinated by FBXL14 through lysines 83 and 106. 293T cells were co-transfected with pMSCV–3× FLAG–3× HA–HES1 or pMSCV–3× FLAG–3× HA-0HES1 (K83A/K106A), pEGFPC2–FBXL14, and pRK5-HA-Ub as indicated for 44 h followed by incubation with MG132 for 4 h, and the cells were harvested for immunoprecipitation (IP) using anti-HES1 antibody under denaturing condition and immunoblotted with anti-HA antibody. Whole-cell extracts (WCE) were immunoblotted with anti-GFP and anti-HA antibodies to indicate expression of exogenous FBXL14 and HES1. The experiment was biologically repeated at least three times.