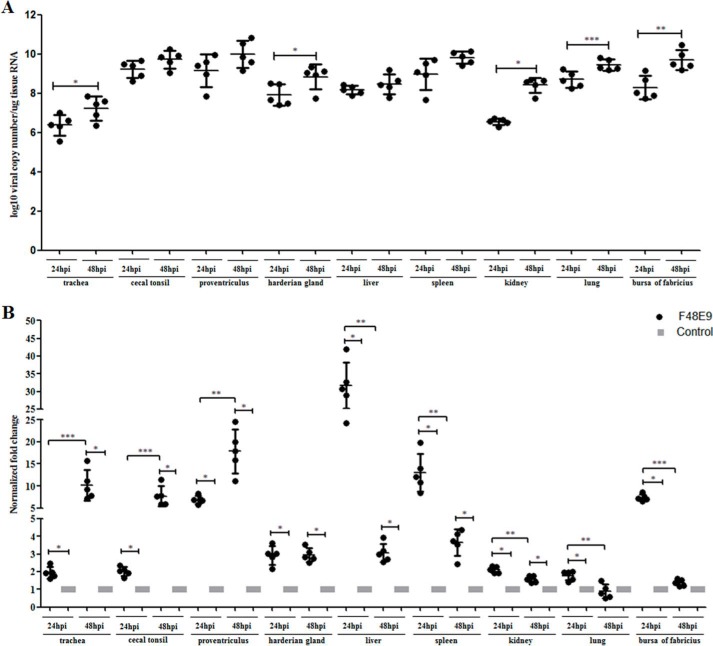
Figure 1.
Viral load and expression of CG-1B mRNA were quantified using real-time RT-PCR. A, viral loads were determined in tissue samples of five NDV F48E9-infected and control birds. The average viral copy number per 1 μg of RNA of each tissue was calculated using a standard curve based on 10-fold dilution series of standard templates with known concentration. No NDV RNA was detected in chickens from the control group. We analyzed the change in viral load in each detected tissue from 24 to 48 hpi. B, transcript alteration of CG-1B gene in NDV F48E9-infected chickens. Comparison of mRNA levels of CG-1B gene at 24 and 48 hpi. Total RNA extracts were prepared from the tissue samples of F48E9-infected and control birds and measured by real-time RT-PCR. Data were normalized with expression of 18S rRNA gene, and mRNA expression of CG-1B gene was calculated relative to that of the control group (relative expression = 1). Change in mRNA level of CG-1B in each tissue at 24 hpi was compared with that of 48 hpi. Data represent means of five biological replicates per group. Error bars indicate S.D. of the mean. Data were compared using the Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.