Abstract
Mitochondrial DNA (mtDNA) is essential for cell viability because it encodes subunits of the respiratory chain complexes. Mitochondrial topoisomerase IB (TOP1MT) facilitates mtDNA replication by removing DNA topological tensions produced during mtDNA transcription, but it appears to be dispensable. To test whether cells lacking TOP1MT have aberrant mtDNA transcription, we performed mitochondrial transcriptome profiling. To that end, we designed and implemented a customized tiling array, which enabled genome-wide, strand-specific, and simultaneous detection of all mitochondrial transcripts. Our technique revealed that Top1mt KO mouse cells process the mitochondrial transcripts normally but that protein-coding mitochondrial transcripts are elevated. Moreover, we found discrete long noncoding RNAs produced by H-strand transcription and encompassing the noncoding regulatory region of mtDNA in human and murine cells and tissues. Of note, these noncoding RNAs were strongly up-regulated in the absence of TOP1MT. In contrast, 7S DNA, produced by mtDNA replication, was reduced in the Top1mt KO cells. We propose that the long noncoding RNA species in the D-loop region are generated by the extension of H-strand transcripts beyond their canonical stop site and that TOP1MT acts as a topological barrier and regulator for mtDNA transcription and D-loop formation.
Keywords: DNA topoisomerase; long noncoding RNA (long ncRNA, lncRNA); mitochondria; mitochondrial DNA (mtDNA); transcription; Top1mt
Introduction
Mitochondrial DNA (mtDNA) is essential because it encodes subunits of the respiratory chain complexes (1). Each mitochondrion contains multiple copies of a 16-kb circular genome (2), and both mtDNA strands, denoted as heavy (H) 3 and light (L), are transcribed. In the mitochondrial genome the only large-size noncoding region (NCR) is confined to a DNA track of ∼700 bp in mice and 1100 bp in humans, which contains the promoters for mtDNA transcription and the origin of H-strand replication (Fig. 1). In addition to the 13 mRNAs coding for the respiratory chain subunits, mtDNA encodes two rRNAs and 22 tRNAs. Ribosomal RNAs associate with nuclear-encoded ribosomal proteins to form the mitochondrial ribosomes, and transfer RNAs are used for mitochondrial protein synthesis.
Figure 1.
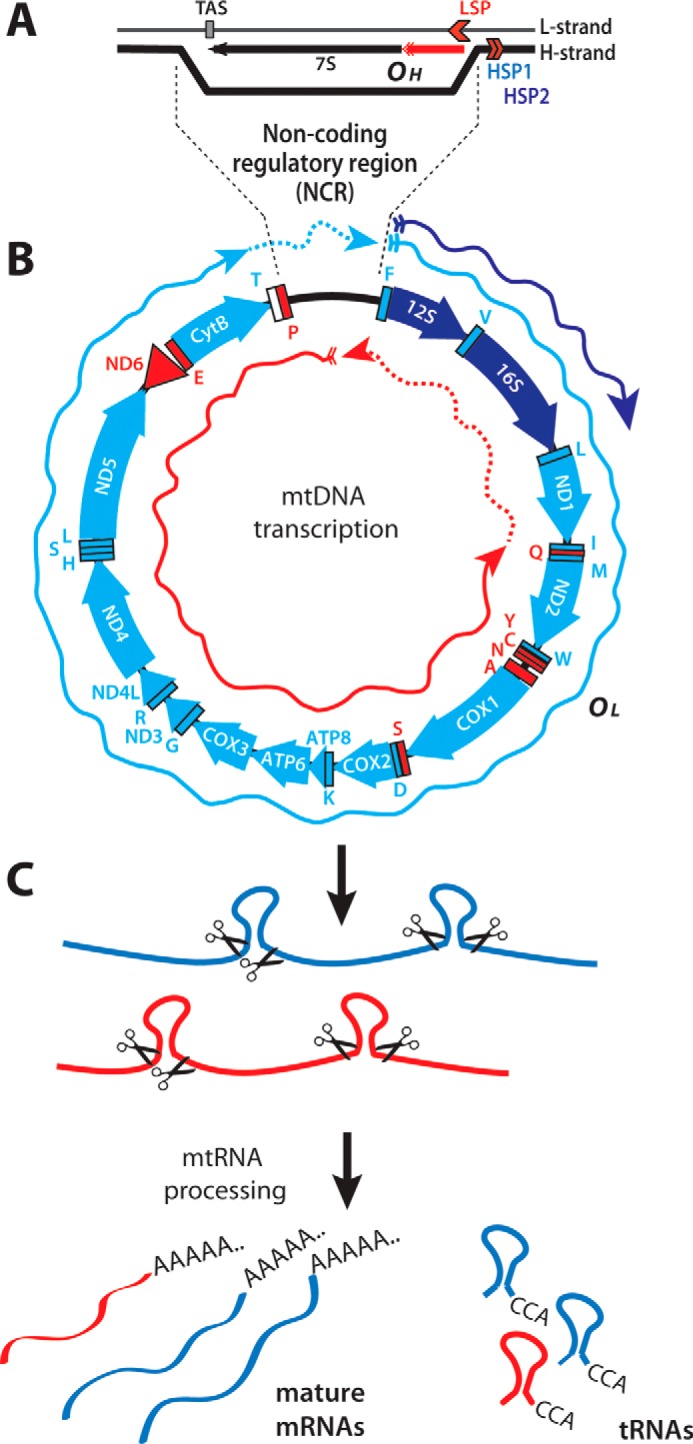
Mammalian mitochondrial genome and its transcripts. A, schematic representation of the noncoding regulatory region containing the D-loop mtDNA formed by the extended RNA primer (red) originating from the L-strand promoter (LSP). TAS, termination-associated sequence. B, the mitochondrial genes are schematized using clockwise and counterclockwise arrows, representing genes encoded by the H- and L-strands, respectively. The H-strand transcripts (rRNAs, mRNAs, and tRNAs) are represented in blue (light or dark blue for products generated from HSP2 or HSP1, respectively). tRNAs and the only mRNA (ND6) produced by L-strand transcription are sketched in red. Blue and red arrows represent the polycistronic transcripts produced by transcription initiated at the H- and L-strand promoters, respectively. C, the polycistronic RNA precursors are processed by excising the tRNAs flanking the open reading frames. Mature messenger and ribosomal RNAs are then maturated by the addition of a 3′-poly(A) tail before being translated or incorporated into mitochondrial ribosomes.
mtDNA transcription starts from three different promoters, two on the H-strand (HSP1 and HSP2) and one on the L-strand (LSP) (3) (Fig. 1A). Transcription starting from HSP2 and LSP produces genome-long polycistronic transcripts, whereas transcription from HSP1 stops prematurely, generating high levels of the two ribosomal RNAs (Fig. 1B). The polycistronic transcripts are processed by the excision of tRNAs, which releases individual mRNAs and rRNAs (4) (Fig. 1C). The tRNAs are further processed by the addition of a CCA triplet at their 3′-end (5), whereas rRNAs and mRNAs mature by the addition of 50–60-nt–long poly(A) tails (6) (Fig. 1C). Poly(A) tails are known to increase mRNA stability for nuclear eukaryotic genes, whereas the same post-transcriptional modification promotes RNA degradation in eubacteria (7). In mitochondria, less is known about the role of polyadenylation (8), and poly(A) tails can have different effects on different transcripts, increasing the stability of some mtRNAs and promoting degradation of others (9).
Bidirectional transcription and replication of mtDNA generates DNA supercoiling (topological stress) in the circular mitochondrial genome, which is anchored to the mitochondrial inner membrane in nucleoids (10–12). The canonical function of topoisomerases is to release helical tensions and decatenate newly replicated DNA molecules (13–17). Four topoisomerases have been identified in mammalian mitochondria: mitochondrial topoisomerase I (TOP1MT) (18), topoisomerase IIIα (TOP3A) (19), and more recently topoisomerases IIα and -β (TOP2A and TOP2B) (20, 21). TOP1MT bears a strong mitochondrial targeting sequence (18) and is the only topoisomerase exclusively targeted to mitochondria (17, 22). Its catalytic core domain is highly similar to the nuclear topoisomerase IB (TOP1), and it is likely that the two enzymes originated from the duplication of a common ancestral gene during vertebrate evolution (22, 23).
TOP1MT has recently been shown to facilitate mtDNA replication during liver regeneration and following ethidium bromide exposure (24). Yet, TOP1MT is not essential for mouse development, probably due to the presence of TOP3A, TOP2A, and TOP2B in mitochondria complementing its function (21). TOP1MT associates with transcriptionally active nucleoids and interacts directly with the mitochondrial RNA polymerase (POLRMT) (25). In addition, ChIP-on-Chip experiments have shown that TOP1MT preferentially binds to mtDNA promoters, suggesting that the enzyme acts as a cofactor for mtDNA transcription. Top1mt KO mouse embryonic fibroblasts (MEFs) show mitochondrial dysfunctions characterized by reduced ATP levels and increased reactive oxygen species production (26). Steady-state levels of mitochondrial transcripts (25, 26) and negative supercoiling of mtDNA are also increased in TOP1MT-deficient cells (21), indicating the role of TOP1MT in removing DNA topological tensions produced by mtDNA transcription.
Here we set out to characterize the mtDNA transcription profile in human and mouse cells and tissues in the absence of TOP1MT. Using customized, strand-specific, tiling arrays, we provide a method to quantify coding and noncoding mitochondrial transcripts on both mtDNA strands simultaneously. This approach offers new insights into the role of TOP1MT as a regulator of mtDNA transcription and replication fork.
Results
Tiling array design for the study of mitochondrial transcripts
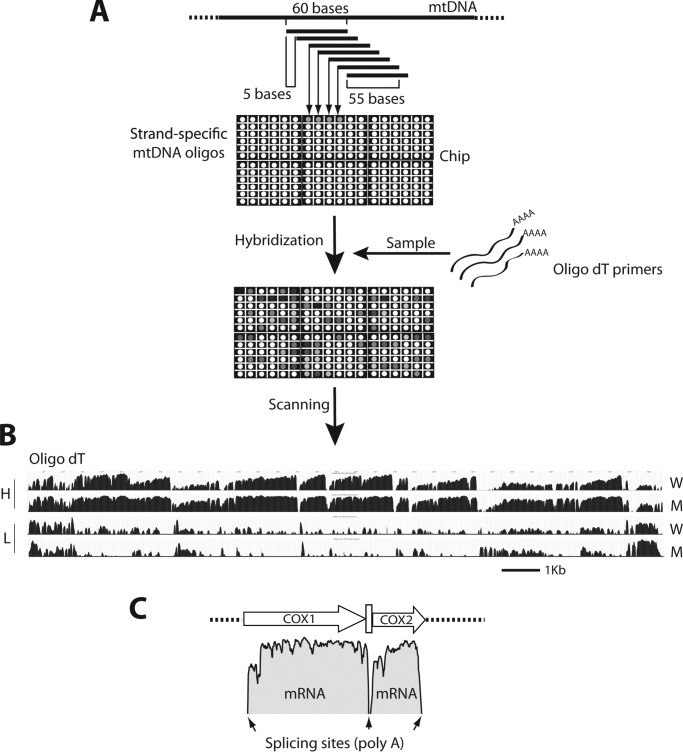
To perform a comprehensive characterization of the mitochondrial transcriptome in the absence of TOP1MT, a non-essential component of the mitochondrial transcription machinery (25), we designed two customized tiling arrays for the mitochondrial genome, one specific for mouse and another for human. Individual probes on the arrays consisted of 60 bases of DNA oligonucleotides that matched the mtDNA sequences and overlapping adjacent probes by 55 nucleotides (Fig. 2A) (sequences are listed in supplemental Table 1). RNA samples were reverse-transcribed and hybridized to the array to generate strand-specific mtDNA transcription profiles. Because our main interest was to analyze mtDNA transcripts (i.e. mtRNA processing and stability), RNA samples were reverse-transcribed using oligo(dT) primers and mature (i.e. polyadenylated) mtRNAs profiles were analyzed.
Figure 2.
Experimental design. A, array organization and experimental protocol. Each probe of the array is a 60-base strand-specific mtDNA sequence overlapping the two adjacent probes by 55 nucleotides. To detect mitochondrial transcripts, RNAs are retrotranscribed using oligo(dT) primers. The cDNA fragments obtained are hybridized to the chip and their abundance measured based on the signal intensity for every probe. B, transcription profiles obtained from the hybridization of mature RNAs on the array. W and M indicate profiles obtained using RNA extracted from whole cells or isolated mitochondria, respectively. H and L refer to heavy or light strand profiles. C, data representation. The intensity of each probe (log scale) is plotted along the mtDNA sequence. Polyadenylated COX1 and COX2 transcripts are shown as examples. Small arrows indicate the location of tRNAs.
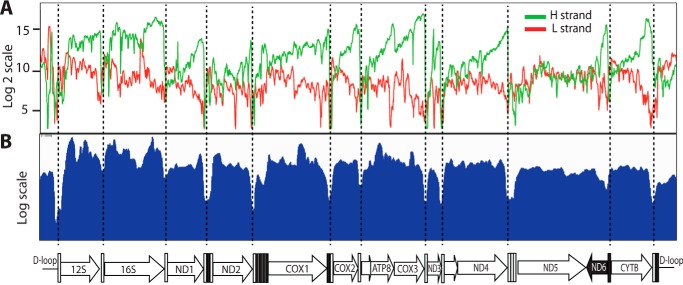
Fig. 2B shows the relative abundance and expression profiles of polyadenylated mtRNAs from WT MEFs. Hybridization of mature RNAs showed a defined pattern characterized by high signals in the coding regions separated by steep drops (“valleys”) at their junctions corresponding to the tRNA processing sites (Fig. 2, B and C). Such a profile is consistent with the mechanism of mtDNA transcript maturation, with mRNAs processing at the tRNA junctions and successive polyadenylation of their 3′-ends (see Fig. 1). The abundance of mature H-strand transcripts was high, consistent with the fact that 12 of the 13 mitochondrial mRNAs and the two mitochondrial rRNAs are transcribed from the H-strand. By contrast, transcripts generated from the L-strand promoter showed much lower signals, consistent with the fact that L-strand transcription codes for only one gene (ND6). A comparison of RNA samples isolated from whole cells (W) or isolated mitochondria (M) showed nearly identical profiles (Fig. 2B). Moreover, the ORF patterns matched the ones generated by standard RNA-seq (Fig. 3), demonstrating the high specificity of the array for mitochondrial transcripts and its effectiveness using total cellular RNA.
Figure 3.
Comparison between mitochondrial transcription profiles of human colon carcinoma HCT116 cells obtained by the tiling array of polyadenylated mtRNAs (A) and RNA-seq (B). Note the similarity between the ORF patterns. Endonucleolytic cleavage at tRNA sites is indicated as vertical dotted lines. mtDNA organization is shown in the bottom panel.
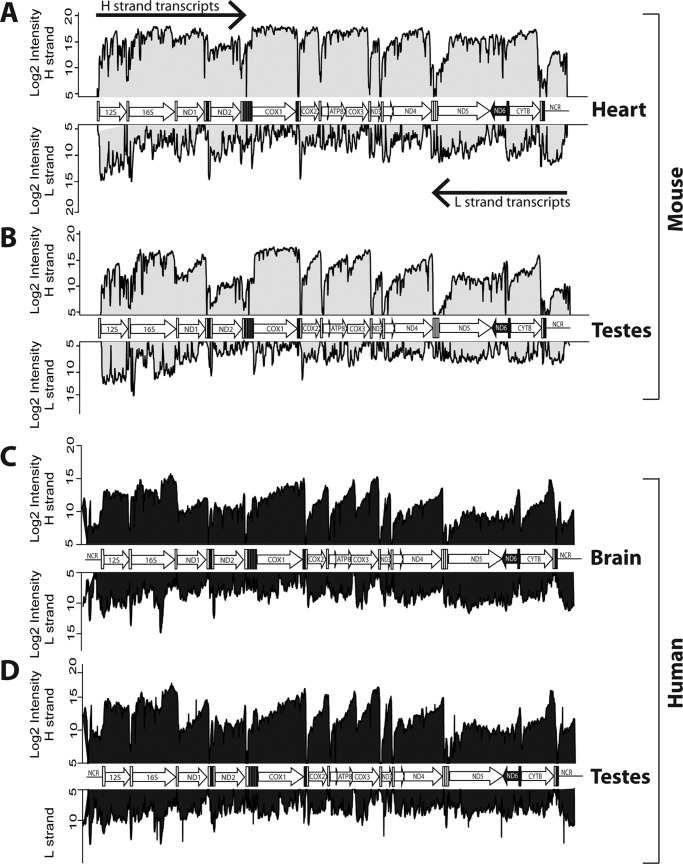
Having established the effectiveness of our tiling array, we explored the mitochondrial transcriptomes of murine and human tissues in parallel. Fig. 4 shows that, in all the tissues analyzed, the H-strand transcripts had consistent and well-defined profiles characterized by high signal blocks within the 12 ORFs separated by deep drops at their junctions, corresponding to the tRNA processing sites. The higher levels toward the 3′-end of each transcript is likely caused by the reverse transcription reaction, which produces a decline in the signals, as the extension was further away from the poly(A) tails. Mitochondrial rRNAs (12S and 16S) also gave high readings, consistent with the fact that both ribosomal and messenger RNAs are processed by polyadenylation (1, 4).
Figure 4.
Polyadenylated mtRNA profiles in mouse heart (A) and testes (B) and human brain (C) and testes (D). The H- and L-strand transcripts are plotted mirrored (top and bottom of each panel, respectively). The organization of the mitochondrial genome is sketched in the middle of each panel.
Signals for the L-strand were uniformly weaker than for the H-strand, reflecting that the L-strand only encodes the ND6 transcript. Unlike the well-defined transcript of the H-stand, the ND6 transcript was not clearly delineated and appeared to include the ND5 region, which is consistent with the lack of a canonical polyadenylation tail at the ND6 3′-end (28). A detailed examination of the L-strand transcriptome also revealed the existence of mature noncoding RNAs (ncRNAs), which were complementary to the H-strand ORFs across different tissues (see Fig. 4, for example the CYTB, COX2, and COX3 regions). These results confirm, via a different and independent method, the presence of antisense noncoding mitochondrial transcripts (29).
Defined signals corresponding to the NCR were also detected on both strands, indicating the presence of noncoding RNAs emanating from both strands of the regulatory NCR. This result reinforces independent observations suggesting the presence of polyadenylation sites within the NCR (28, 30). The signals in the NCR were clearly reproducible across tissues and were detectable both in murine and human samples.
Transcription profile of Top1mt knock-out cells reveals increased noncoding RNAs in the mtDNA noncoding regulatory region
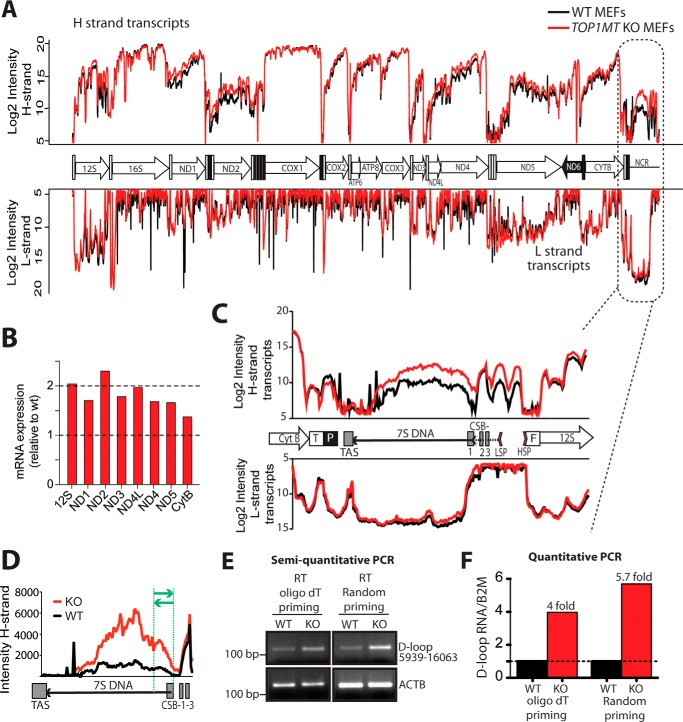
Based on the association of TOP1MT with the mitochondrial transcription machinery including POLRMT (25), and the fact that inactivation of nuclear TOP1 selectively impacts the expression of nuclear genes (31–33), we compared the mitochondrial transcriptome profiles of MEFs from KO and WT mice (21, 26). Mitochondrial transcription patterns were globally similar (Fig. 5A). Yet, quantification of the signal intensity within different ORFs showed that mitochondrial transcripts were ∼1.5–2-fold more abundant in Top1mt KO MEFs compared with WT MEFs (Fig. 5B). This quantitative difference is consistent with quantitative RT-PCR results (25, 26), demonstrating that TOP1MT does not affect the processing of mitochondrial transcripts.
Figure 5.
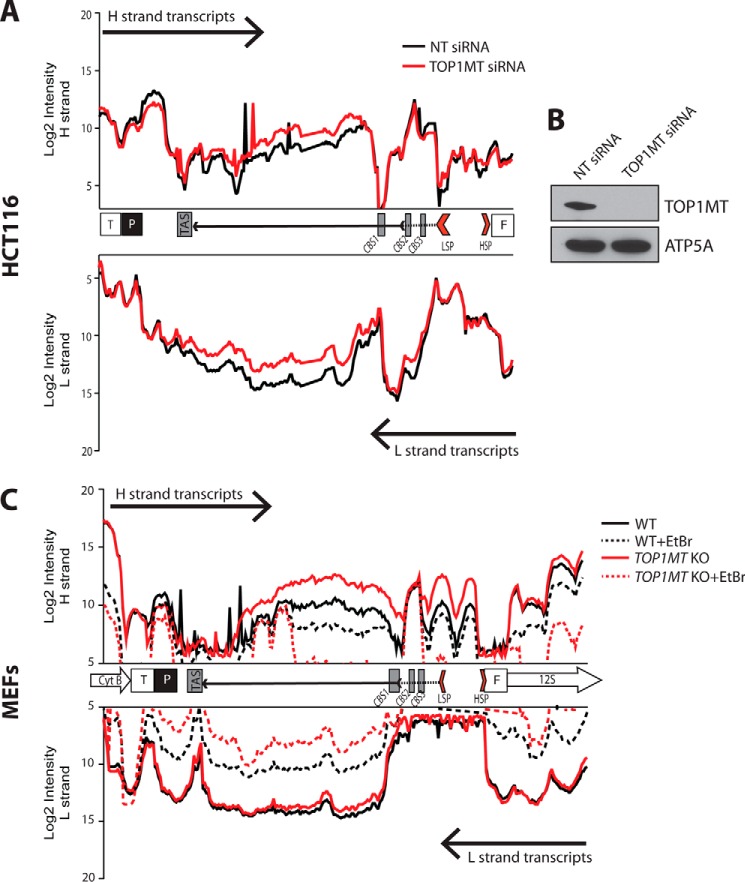
Differential mtRNA profiles in the regulatory region of WT and Top1mt KO MEFs. A, comparison of the mtDNA transcription profiles in WT and KO MEFs (black and red, respectively). B, quantification of mitochondrial transcripts in Top1mt KO MEFs. The average intensities of the probes within different ORFs were added together, and the values relative to WT for each gene were plotted. C, polyadenylated transcripts detected in the mtDNA NCR. The NCR organization is schematized between the H- and L-strand transcription profiles. Regulatory sequences within the NCR are abbreviated as HSP and LSP (H- and L-strand promoters, respectively), TAS (termination-associated sequence), and CSB-1, −2, and -3 (conserved sequence boxes 1, 2, and 3). D, quantitation of the abundance of the noncoding RNA transcript produced by H-strand transcription and spanning the 7S DNA. Intensities for each probe of the tiling array are represented as a linear scale. E and F, quantitation of the ncRNA by semiquantitative and quantitative RT-PCR. Total RNA was retrotranscribed with oligo(dT) or random primers. The cDNA was used as a template to amplify mtDNA nucleotides 15939–16063. The region of the noncoding RNA amplified by PCR is shown shown between the green lines in D. For semiquantitative PCR, PCR products were run on 2% agarose gels, and the band intensities were visualized following UV transillumination. β-Actin (ACTB) was co-amplified as a control. For quantitative PCR, the amounts of PCR products in Top1mt KO samples were quantified using the ΔΔCt method and are expressed relative to the WT.
In addition, a comparison between the transcript profiles of WT and Top1mt KO MEFs showed selective up-regulation of noncoding RNAs spanning the region of the 7S DNA and corresponding to the end of the H-strand transcription (Fig. 5C). The levels of these polyadenylated RNAs were about 5-fold higher in the Top1mt KO MEFs compared with WT MEFs (Fig. 5D). To validate this finding, we performed RT-PCR experiments and quantified RNA species by semiquantitative and quantitative RT-PCR (Fig. 5, E and F, respectively). Both methods confirmed the increase of the noncoding RNA in Top1mt KO MEFs, and the magnitude of this increase matched the 5-fold changes determined by the tiling array.
To support the causal link between TOP1MT and the presence of noncoding RNA transcripts corresponding to the end of the H-strand transcripts in the regulatory region of the mitochondrial genome, we performed TOP1MT knockdown experiments in human colon carcinoma HCT116 cells (Fig. 6, A and B). Consistent with the results in the murine Top1mt KO cells, we found that acute TOP1MT silencing strongly increased the levels of the H-strand ncRNA spanning the D-loop region.
Figure 6.
ncRNA in the regulatory region of WT and Top1mt KO MEFs. A, polyadenylated transcripts detected in the mtDNA noncoding region of human HCT116 cells after acute TOP1MT knockdown. B, Western blotting showing the efficiency of TOP1MT knockdown at 72 h after siRNA transfection. ATP5A served as a control for equal loading. C, stability of the regulatory region ncRNAs in WT (black) and Top1mt KO MEFs (red). The stability of polyadenylated ncRNAs was assessed by blocking mtDNA transcription with EtBr. The abundance of polyadenylated mtRNAs before and after treatment with EtBr is represented by the solid and dotted lines, respectively. TAS, termination-associated sequence. T, threonine tRNA; P, proline tRNA; F, phenylalanine tRNA.
The increase of the ncRNA species produced by H-strand transcription, occurring in the mtDNA regulatory region in the absence in TOP1MT, could be due to enhanced stability or, alternatively, to an increase in transcription rate. To understand the basis for this strong up-regulation, we treated WT and Top1mt KO MEFs with ethidium bromide (EtBr). EtBr is a high-affinity DNA intercalating dye commonly used to inhibit mtDNA transcription and replication. Due to the relatively long half-life of mtDNA (34), a short treatment with EtBr effectively blocks mtDNA transcription without affecting the mtDNA copy number. EtBr treatment caused a marked decrease in the ncRNA levels of the mtDNA regulatory region (Fig. 6C). This decrease was markedly more pronounced in Top1mt KO MEFs, suggesting a shorter half-life of these ncRNA species. These data suggest that the increase in ncRNA in the mtDNA regulatory region is due to enhanced transcription.
7S DNA alterations in Top1mt KO MEFs
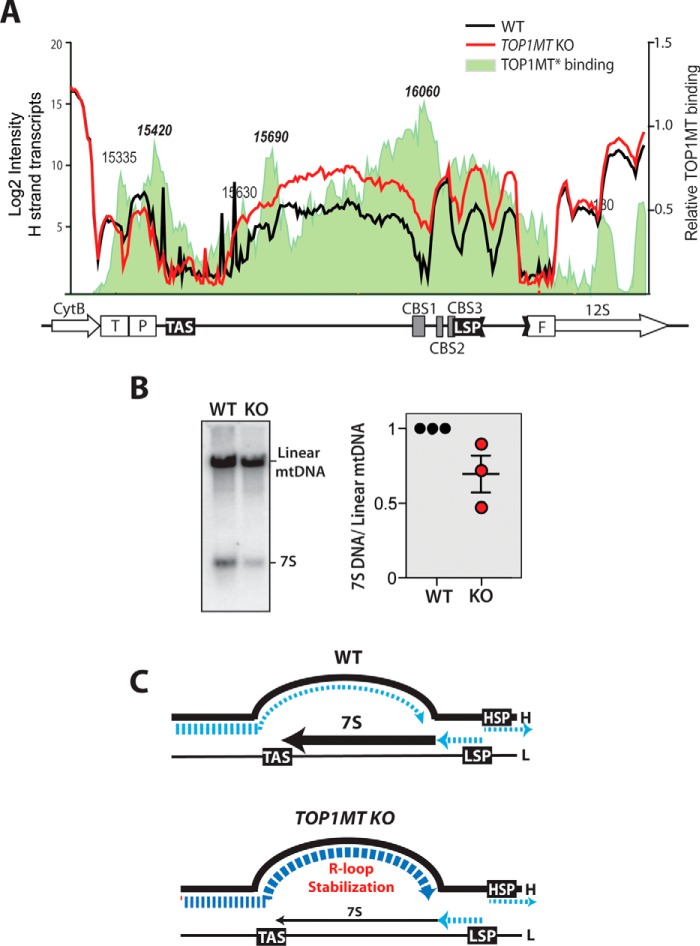
TOP1MT forms high-affinity complexes in the noncoding region of mtDNA (35, 36) (Fig. 7A), and a lack of TOP1MT leads to profound changes in the topological state of the mitochondrial genome, resulting in more negatively supercoiled mtDNA molecules (19). The noncoding RNA, which we found to be strongly enhanced in the absence of TOP1MT, is produced by H-strand transcription; thus, we hypothesized that the topological changes occurring in the NCR upon TOP1MT loss facilitate the progression of H-strand transcription into this region, generating high levels of ncRNAs. We hypothesized that increased-H strand transcription into the NCR could interfere with the production of 7S DNA, which is synthetized by DNA polymerase γ in the opposite direction. Indeed, we found that 7S DNA levels were decreased in Top1mt KO MEFs (Fig. 7B). These results suggest that TOP1MT might play a previously unknown role in the mtDNA NCR by regulating the termination of transcription and facilitating replication (21, 24, 37).
Figure 7.
Regulatory role of TOP1MT on transcription and replication of the NCR. A, alterations of the NCR transcription patterns in Top1mt KO cells in relation to TOP1MT binding sites. TOP1MT binding sites are represented by the shaded area colored in light green and were determined using the tiling array after isolation of TOP1MT cleavage complexes (see “Experimental procedures”). B, Southern blot analysis of 7S DNA in WT and Top1mt KO cells. The right panel shows results from three independent determinations. C, schematic representation of the NCR transcript and 7S aberrations in Top1mt KO MEFs. DNA and RNA species are represented in black and blue, respectively (see “Discussion” for details).
Discussion
To investigate the impact of TOP1MT depletion on the mitochondrial transcriptome, we designed customized tiling arrays allowing simultaneous quantitative and qualitative analysis of all mitochondrial transcripts. Although a comprehensive map of the mitochondrial transcriptome has been generated recently using RNA-seq (30), RNA-seq requires specialized bioinformatics support to analyze the results. The advantage of the tiling array technique is its simplicity and strand specificity, even when using total RNAs as the starting material (see Fig. 2). A comparison between the mitochondrial transcription profiles generated using the tiling array or the RNA-Seq approach showed similar patterns (see Fig. 3). The mtDNA open reading frames were clearly detectable with both techniques and their profiles were comparable, although the RNA-Seq data reflected the combined transcript for both mtDNA strands. The ncRNAs spanning the mtDNA noncoding region were also readily detected in high abundance by RNA-Seq. Moreover, the relative abundance of the different coding and noncoding transcripts was comparable across the two different techniques. Thus, our tiling array method enables the study of mtDNA transcription features under different experimental conditions without purifying mitochondria.
RNA processing and maturation represent key features of gene expression regulation. In mitochondria, newly synthetized transcripts, produced as long polycistronic units, are polyadenylated at the 3′-end by the mitochondrial poly(A) polymerase (38). Contrary to the nuclear compartment where polyadenylation stabilizes mRNAs, the function of poly(A) tails in mitochondria appears to differ (7), as polyadenylation has opposite effects on the stability of different mitochondrial transcripts (9).
Here we set out to study mtDNA gene expression by profiling all of the polyadenylated (i.e. mature) mtRNAs in mouse and human samples under physiological conditions in the presence of EtBr and in TOP1MT-deficient cells. The expression profiles obtained for both the mouse and human genomes were similar and consistent with the tRNA punctuation model for mtRNAs processing (4). On the H-strand, the regions corresponding to ribosomal and messenger RNAs gave high readings, exactly matching the 5′- and 3′-ends of each transcript. The products of the L-strand transcription showed much lower signals, which is consistent with the fact that the L-strand contains only one ORF for ND6. Notably, ND6 transcripts were barely detectable in both human and mouse samples (Figs. 3 and 4). This result confirms previous data showing that the ND6 transcript is polyadenylated ∼500 nucleotides downstream of its stop codon (28). Accordingly, we picked up a clear signal in the region complementary to ND5, which could represent the 3′-end of the processed ND6 transcript.
Low-abundance polyadenylated L-strand transcripts were clearly detectable outside of the ND6 region both in murine and human tissues (Figs. 4 and 5). These ncRNAs tended to show the same valleys (tRNA punctuation) as those from the H-strand and matched the antisense sequence of many H-strand mRNAs (see Figs. 3–5, the CYTB, ND5, ND4, COX3, COX2 and ND1 regions). The existence of three long noncoding mitochondrial RNAs complementary to ND5, ND6, and CYTB has been described by Rackham et al. (39). Here we confirmed the presence of these three antisense RNAs using an independent method, and additionally, we have shown that their abundance is variable, generally being fairly high in human solid tissues and lower in MEFs (Figs. 3 and 4). Similar to nuclear antisense RNAs (40), these mitochondrial RNA species may be involved in the regulation of mtDNA gene expression, and their production could depend on different metabolic requirements in different cellular types (29).
In addition to the antisense ncRNAs, we report the existence of ncRNAs mapping to the mitochondrial regulatory region (NCR). Noncoding RNAs complementary to 7S DNA have been reported recently in the mitochondrial genome (16). These RNAs form an R-loop on the mtDNA regulatory region and play a role in mtDNA organization and mitochondrial dysfunction.
The functions of TOP1MT have remained relatively unexplored since the discovery of the gene 15 years ago (18, 22). In contrast to other nuclear-encoded genes that sustain mtDNA maintenance, TOP1MT is not essential (21, 24, 26, 37) and is restricted to vertebrates (22). Two recent studies show that TOP1MT is required for mtDNA copy number expansion during liver regeneration (24) and myocardial adaptation to mitochondrial poisoning by doxorubicin (37). TOP1MT also tends to repress mitochondrial transcription (25). Consistently, our genome-wide analyses showed increased steady-state levels of most mitochondrial mRNAs in the absence of TOP1MT, a result that confirms previous quantitative RT-PCR data (26). Yet, the overall transcriptome profiles were remarkably similar, indicating that TOP1MT is not critical for mtDNA processing. Interestingly, the most altered transcripts in Top1mt-deficient MEFs were the noncoding RNAs produced by the H-strand transcription and spanning the regulatory region of the D-loop. A noncoding RNA encoded by this region has been reported by other groups using different techniques and model systems (28, 30, 41–43), but its function and regulation remains unclear. We found that, in cells lacking TOP1MT, such noncoding RNA complementary to the 7S DNA was 5-fold enriched, representing a notable perturbation of the entire mtDNA expression profile (Figs. 5–7). EtBr treatment strongly down-regulated this RNA in the Top1mt KO MEFs, whereas it had only a partial effect in the WT MEFs (Fig. 6). This result indicates that the up-regulation of this RNA species in Top1mt KO MEFs is not due to an increase in its stability.
To explain our finding that TOP1MT attenuates the expression of the noncoding RNA mapping the NCR, we propose that TOP1MT, tuning mtDNA topology, limits H-strand transcription into the mtDNA regulatory region (see model in Fig. 7C). Accordingly, TOP1MT cleavage sites accumulate in the mtDNA D-loop region (35) (Fig. 7A). Moreover, the mtDNA of Top1mt KO MEFs is highly negatively supercoiled (i.e. underwound) (21), which could enable transcription progression behind its usual termination point. In line with this hypothesis, we found decreased 7S DNA (produced in the opposite direction) in Top1mt KO MEFs. Hence, we surmise that TOP1MT acts as a regulator of mtDNA topology in NCR by facilitating 7S formation and termination of H-strand transcription at the termination-associated sequence (44). These data suggest a novel regulatory function for TOP1MT as an enzyme able to modulate mtDNA transcription through modification of mtDNA topology.
Experimental procedures
Cell culture and TOP1MT knockdown
Human HCT116 cells and WT and Top1mt KO MEFs (26) were cultured in DMEM supplemented with 10% FCS. To inhibit mitochondrial transcription, WT and Top1mt KO MEFs were treated with 50 ng/ml ethidium bromide for 24 h prior to RNA extraction. For TOP1MT knockdown, HCT116 cells were transfected with 5 nm TOP1MT siRNA (ON-TARGETplus SMARTpool, Dharmacon) or scrambled sequences using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's specifications. Knockdown efficiency was tested by Western blot analysis 72 h after transfection.
RNA isolation and mitochondrial DNA tiling array
Total RNA was isolated from cells (2 × 106) using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. For RNA isolation from mice tissues, the brain and testes were homogenized in 3 ml of TRIzol using an Ultra-Turrax homogenizer. After the addition of 600 μl of chloroform, samples were thoroughly vortexed and spun at 12,000 × g for 15 min at 4 °C. The aqueous phase was then precipitated with equal volumes of RNase-free 70% ethanol and loaded onto the RNeasy mini kit columns (Qiagen). Washing and RNA elution were performed according to the manufacturer's instructions. For all extractions, DNase digestion was performed on the columns. Human total RNA was purchased from Agilent Technologies (Santa Clara, CA).
Arrays were manufactured by Agilent Technologies using Agilent SurePrint 4 × 44K format. Tiling used 60-mer probes with 5bp stagger for both mtDNA strands (H and L) to detect strand-specific expression within the same array. The arrays also included probes for nuclear genes that served as loading controls. Tiling arrays were customized for the human and murine mitochondrial genomes.
Total RNA isolated from cultured cells or tissues was labeled with the Quick Amp labeling kit (Agilent, catalog no. 5190-0442) following the manufacturer's recommended protocols. Briefly, cRNA was generated from 500 ng of RNA with T7 promoter-tagged oligo(dT) and Moloney murine leukemia virus (MMLV) reverse transcriptase. cRNA was labeled with Cy3 and Cy5 dye using T7 RNA polymerase. Three μg of labeled cRNA was fragmented and hybridized to the array at 65 °C for 17 h. Slides were washed in Agilent Gene Expression wash buffer 1 for 1 min at room temperature and in wash buffer 2 for 1 min at 37 °C. The slides were scanned in an Agilent C-scanner and the data extracted with Agilent Feature Extraction software 10.7.
RNA-Seq analysis
RNA was isolated from HCT116 cells as described above. The sequencing libraries were generated using the TotalScriptTM RNA-Seq kit (Epicenter) and sequenced using the HiSeq 2000 (Illumina) with paired end 100-bp reads. Data were visualized by alignment to the human genome assembly 19.
Semiquantitative and quantitative PCR
The relative abundance of the noncoding RNA spanning the D-loop region was assessed using semiquantitative and quantitative PCR. For semiquantitative measurement, total RNA was retrotranscribed with oligo(dT) or random primers, and cDNA served as the template for the amplification of the D-loop portion spanning nucleotides 15645 to 15812. Actin B was amplified as a reference. To ensure that the abundance of PCR product and template was proportional, we limited the amplification step to 18 cycles. The primers used were the following: D-loop forward, GGAAGGGGATAGTCATATGGAAGAGAAG; D-loop reverse, TGGCCCTGAAGTAAGAACCAGATGTCTGATAAAG; ACTB forward, CTACGAGGGCTATGCTC; and ACTB reverse, CTTTGATGTCACGCACG. For quantitative PCR, the PCR reactions were performed in triplicates on 384-well reaction plates (Applied Biosystems). Each PCR reaction (final volume, 10 μl) contained 25 ng of cDNA, 5 μl of Power SYBR-Green PCR Master Mix (Applied Biosystems), and each forward and reverse primer at 0.5 μm. The D-loop region and the normalizing control, actin B, were amplified using the primers listed above.
Analysis of 7S DNA
Southern blot analysis of mtDNA was carried out as described (27). Briefly, 2 μg of total DNA was digested with the mtDNA single-cutter XhoI and run on a 1% agarose gel. DNA was then transferred by capillary transfer onto Hybond N+ membrane (GE Healthcare) and hybridized with a 32P-labeled probe for mouse-mtDNA (nt 15007–15805). 32P signals were visualized with a PhosphorImager (Molecular Dynamics).
Author contributions
I. D., H. Z., X. W., and Y. P. designed the experiments and analyzed the data. I. D., H. Z., S. K., and X. W. performed the experiments. I. D., H. Z., and Y. P. wrote the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Intramural Research Program Grant Z01 BC006161 from the Center for Cancer Research, NCI. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table 1.
- H
- heavy
- L
- light
- HSP
- H-strand promoter
- LSP
- L-strand promoter
- NCR
- noncoding region
- MEF
- mouse (murine) embryonic fibroblast
- ncRNA
- noncoding RNA.
References
- 1. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 2. D'Erchia A. M., Atlante A., Gadaleta G., Pavesi G., Chiara M., De Virgilio C., Manzari C., Mastropasqua F., Prazzoli G. M., Picardi E., Gissi C., Horner D., Reyes A., Sbisà E., Tullo A., and Pesole G. (2015) Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion 20, 13–21 [DOI] [PubMed] [Google Scholar]
- 3. Bonawitz N. D., Clayton D. A., and Shadel G. S. (2006) Initiation and beyond: Multiple functions of the human mitochondrial transcription machinery. Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 4. Ojala D., Montoya J., and Attardi G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 [DOI] [PubMed] [Google Scholar]
- 5. Levinger L., Mörl M., and Florentz C. (2004) Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 32, 5430–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Montoya J., Ojala D., and Attardi G. (1981) Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290, 465–470 [DOI] [PubMed] [Google Scholar]
- 7. Schuster G., and Stern D. (2009) RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol. Biol. Transl. Sci. 85, 393–422 [DOI] [PubMed] [Google Scholar]
- 8. Bobrowicz A. J., Lightowlers R. N., and Chrzanowska-Lightowlers Z. (2008) Polyadenylation and degradation of mRNA in mammalian mitochondria: A missing link? Biochem. Soc. Trans. 36, 517–519 [DOI] [PubMed] [Google Scholar]
- 9. Wilson W. C., Hornig-Do H. T., Bruni F., Chang J. H., Jourdain A. A., Martinou J. C., Falkenberg M., Spåhr H., Larsson N. G., Lewis R. J., Hewitt L., Baslé A., Cross H. E., Tong L., Lebel R. R., et al. (2014) A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Hum. Mol. Genet. 23, 6345–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kukat C., Davies K. M., Wurm C. A., Spåhr H., Bonekamp N. A., Kühl I., Joos F., Polosa P. L., Park C. B., Posse V., Falkenberg M., Jakobs S., Kühlbrandt W., and Larsson N. G. (2015) Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 112, 11288–11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He J., Mao C. C., Reyes A., Sembongi H., Di Re M., Granycome C., Clippingdale A. B., Fearnley I. M., Harbour M., Robinson A. J., Reichelt S., Spelbrink J. N., Walker J. E., and Holt I. J. (2007) The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 176, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bogenhagen D. F. (2012) Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta 1819, 914–920 [DOI] [PubMed] [Google Scholar]
- 13. Champoux J. J. (2001) DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 14. Wang J. C. (2009) A journey in the world of DNA rings and beyond. Annu. Rev. Biochem. 78, 31–54 [DOI] [PubMed] [Google Scholar]
- 15. Pommier Y., Leo E., Zhang H., and Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akman G., Desai R., Bailey L. J., Yasukawa T., Dalla Rosa I., Durigon R., Holmes J. B., Moss C. F., Mennuni M., Houlden H., Crouch R. J., Hanna M. G., Pitceathly R. D., Spinazzola A., and Holt I. J. (2016) Pathological ribonuclease H1 causes R-loop depletion and aberrant DNA segregation in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, E4276–E4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pommier Y., Sun Y., Huang S. N., and Nitiss J. L. (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Barceló J. M., Lee B., Kohlhagen G., Zimonjic D. B., Popescu N. C., and Pommier Y. (2001) Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 98, 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Lyu Y. L., and Wang J. C. (2002) Dual localization of human DNA topoisomerase IIIα to mitochondria and nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Low R. L., Orton S., and Friedman D. B. (2003) A truncated form of DNA topoisomerase IIβ associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem. 270, 4173–4186 [DOI] [PubMed] [Google Scholar]
- 21. Zhang H., Zhang Y. W., Yasukawa T., Dalla Rosa I., Khiati S., and Pommier Y. (2014) Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIα and IIβ in vertebrate mitochondria. Nucleic Acids Res. 42, 7259–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H., Meng L. H., Zimonjic D. B., Popescu N. C., and Pommier Y. (2004) Thirteen-exon-motif signature for vertebrate nuclear and mitochondrial type IB topoisomerases. Nucleic Acids Res. 32, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dalla Rosa I., Goffart S., Wurm M., Wiek C., Essmann F., Sobek S., Schroeder P., Zhang H., Krutmann J., Hanenberg H., Schulze-Osthoff K., Mielke C., Pommier Y., Boege F., and Christensen M. O. (2009) Adaptation of topoisomerase I paralogs to nuclear and mitochondrial DNA. Nucleic Acids Res. 37, 6414–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khiati S., Baechler S. A., Factor V. M., Zhang H., Huang S. Y., Dalla Rosa I., Sourbier C., Neckers L., Thorgeirsson S. S., and Pommier Y. (2015) Lack of mitochondrial topoisomerase I (TOP1mt) impairs liver regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 11282–11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sobek S., Dalla Rosa I., Pommier Y., Bornholz B., Kalfalah F., Zhang H., Wiesner R. J., von Kleist-Retzow J. C., Hillebrand F., Schaal H., Mielke C., Christensen M. O., and Boege F. (2013) Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res. 41, 9848–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Douarre C., Sourbier C., Dalla Rosa I., Brata Das B., Redon C. E., Zhang H., Neckers L., and Pommier Y. (2012) Mitochondrial topoisomerase I is critical for mitochondrial integrity and cellular energy metabolism. PLoS ONE 7, e41094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruhanen H., Borrie S., Szabadkai G., Tyynismaa H., Jones A. W., Kang D., Taanman J. W., and Yasukawa T. (2010) Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim. Biophys. Acta 1803, 931–939 [DOI] [PubMed] [Google Scholar]
- 28. Slomovic S., Laufer D., Geiger D., and Schuster G. (2005) Polyadenylation and degradation of human mitochondrial RNA: The prokaryotic past leaves its mark. Mol. Cell. Biol. 25, 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dietrich A., Wallet C., Iqbal R. K., Gualberto J. M., and Lotfi F. (2015) Organellar non-coding RNAs: Emerging regulation mechanisms. Biochimie 117, 48–62 [DOI] [PubMed] [Google Scholar]
- 30. Mercer T. R., Neph S., Dinger M. E., Crawford J., Smith M. A., Shearwood A. M., Haugen E., Bracken C. P., Rackham O., Stamatoyannopoulos J. A., Filipovska A., and Mattick J. S. (2011) The human mitochondrial transcriptome. Cell 146, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miao Z. H., Player A., Shankavaram U., Wang Y. H., Zimonjic D. B., Lorenzi P. L., Liao Z. Y., Liu H., Shimura T., Zhang H. L., Meng L. H., Zhang Y. W., Kawasaki E. S., Popescu N. C., Aladjem M. I., et al. (2007) Nonclassic functions of human Topoisomerase I: Genome-wide and pharmacologic analyses. Cancer Res. 67, 8752–8761 [DOI] [PubMed] [Google Scholar]
- 32. Solier S., Ryan M. C., Martin S. E., Varma S., Kohn K. W., Liu H., Zeeberg B. R., and Pommier Y. (2013) Transcription poisoning by topoisomerase I is controlled by gene length, splice sites, and miR-142–3p. Cancer Res. 73, 4830–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. King I. F., Yandava C. N., Mabb A. M., Hsiao J. S., Huang H. S., Pearson B. L., Calabrese J. M., Starmer J., Parker J. S., Magnuson T., Chamberlain S. J., Philpot B. D., and Zylka M. J. (2013) Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gross N. J., Getz G. S., and Rabinowitz M. (1969) Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J. Biol. Chem. 244, 1552–1562 [PubMed] [Google Scholar]
- 35. Dalla Rosa I., Huang S. Y., Agama K., Khiati S., Zhang H., and Pommier Y. (2014) Mapping topoisomerase sites in mitochondrial DNA with a poisonous mitochondrial topoisomerase I (Top1mt). J. Biol. Chem. 289, 18595–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang H., and Pommier Y. (2008) Mitochondrial topoisomerase I sites in the regulatory D-loop region of mitochondrial DNA. Biochemistry 47, 11196–11203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khiati S., Dalla Rosa I., Sourbier C., Ma X., Rao V. A., Neckers L. M., Zhang H., and Pommier Y. (2014) Mitochondrial topoisomerase I (top1mt) is a novel limiting factor of doxorubicin cardiotoxicity. Clin. Cancer Res. 20, 4873–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomecki R., Dmochowska A., Gewartowski K., Dziembowski A., and Stepien P. P. (2004) Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 32, 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rackham O., Shearwood A. M., Mercer T. R., Davies S. M., Mattick J. S., and Filipovska A. (2011) Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C. C., Suzuki M., Kawai J., Suzuki H., Carninci P., Hayashizaki Y., Wells C., Frith M., Ravasi T., Pang K. C., Hallinan J., Mattick J., Hume D. A., Lipovich L., Batalov S., Engström P. G., Mizuno Y., Faghihi M. A., Sandelin A., Chalk A. M., Mottagui-Tabar S., Liang Z., Lenhard B., Wahlestedt C., RIKEN Genome Exploration Research Group, Genome Science Group (Genome Network Project Core Group), and FANTOM Consortium (2005) Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 [DOI] [PubMed] [Google Scholar]
- 41. Freyer C., Park C. B., Ekstrand M. I., Shi Y., Khvorostova J., Wibom R., Falkenberg M., Gustafsson C. M., and Larsson N. G. (2010) Maintenance of respiratory chain function in mouse hearts with severely impaired mtDNA transcription. Nucleic Acids Res. 38, 6577–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sbisà E., Nardelli M., Tanzariello F., Tullo A., and Saccone C. (1990) The complete and symmetric transcription of the main noncoding region of rat mitochondrial genome: In vivo mapping of heavy and light transcripts. Curr. Genet. 17, 247–253 [DOI] [PubMed] [Google Scholar]
- 43. Selwood S. P., McGregor A., Lightowlers R. N., and Chrzanowska-Lightowlers Z. M. (2001) Inhibition of mitochondrial protein synthesis promotes autonomous regulation of mtDNA expression and generation of a new mitochondrial RNA species. FEBS Lett. 494, 186–191 [DOI] [PubMed] [Google Scholar]
- 44. Doda J. N., Wright C. T., and Clayton D. A. (1981) Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl. Acad. Sci. U.S.A. 78, 6116–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.