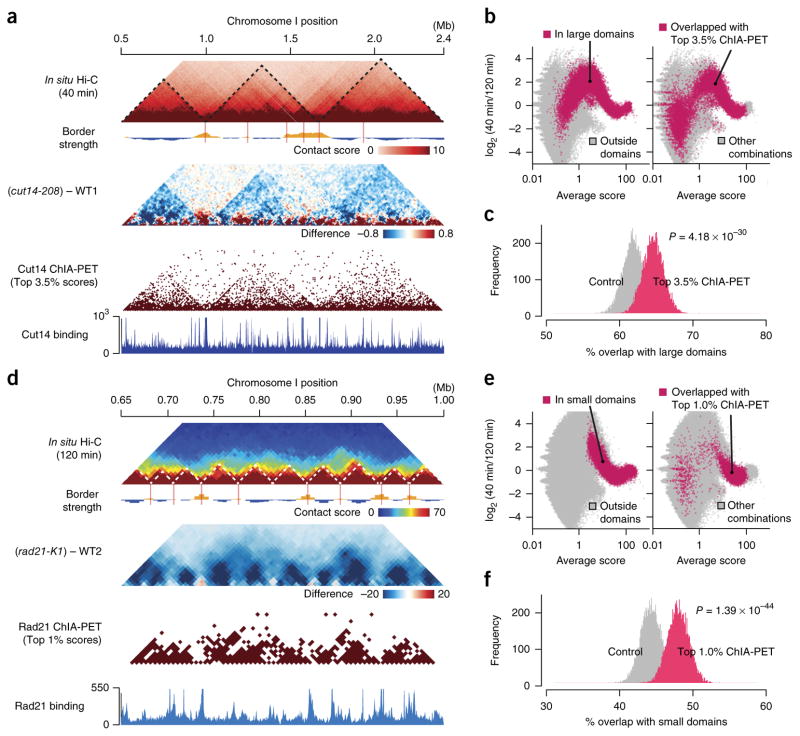
Figure 6.
Comparison between the in situ Hi-C and ChIA-PET data (a) In situ Hi-C data were compared to the Cut14 condensin ChIA-PET data21. Maps for the 1.9-Mb region show contact scores at the 40-min time point (mid M; top) and difference of contact scores between the cut14-208 mutant and WT1 (second). The ChIA-PET data were analyzed using the same procedure applied to the in situ Hi-C data (Supplementary Fig. 1b). The top 3.5% of condensin-mediated contacts were extracted from the ChIA-PET data and plotted (third). Based on the ChIA-PET data, condensin enrichment and binding sites were predicted (bottom) as described in ref. 21. (b) Averages and log2 ratios of contact scores between the 40- and 120-min points were plotted for genomic contacts within (red) and outside (gray) the large domains (left). The same data were annotated differently for genomic contacts that overlapped with the top 3.5% condensin-mediated contacts (red) and others (gray) (right). (c) The top 3.5% of condensin-mediated contacts predicted from the ChIA-PET data were frequently positioned within the large domains (Online Methods). 1,000 genomic contacts were randomly selected from the top 3.5% of condensin ChIP-PET combinations. Genomic contacts with the same distances were randomly selected for distance-conserved random sampling from every combination. This sampling was repeated 10,000 times, and distributions of % overlap with the large domains were plotted. For P value calculation, the same sampling was repeated 100 times, and distributions of % overlap scores were subjected to one-sided Student’s t test. (d) The in situ Hi-C data were compared to the Rad21 cohesin ChIA-PET data21. Based on the ChIA-PET data, cohesin enrichment and binding sites were predicted (bottom) as described in ref. 21. (e) Averages and log2 ratios of contact scores between the 40- and 120-min time points were plotted for genomic contacts within (red) and outside (gray) the small domains (left). The same data were differently annotated for genomic contacts that overlapped with the top 1% of cohesin-mediated contacts (red) and others (gray) (right). (f) The top 1% of cohesin-mediated contacts predicted from the ChIA-PET data were often located within the small domains. The same analysis as described in c was performed for the cohesin-mediated contacts and the small domains.