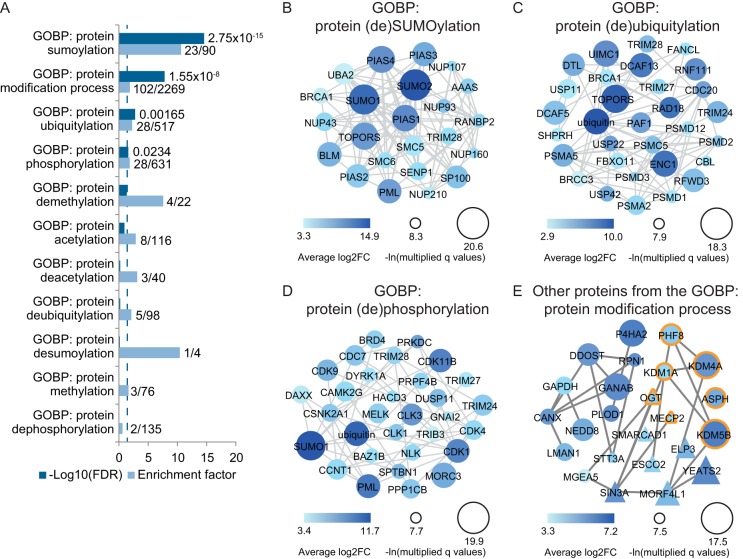
Fig. 3.
Detailed analysis of the co-modified targets reveals extensive potential crosstalk among PTMs. A, Significantly enriched co-modified targets upon inhibition of the proteasome were annotated and a gene ontology enrichment analysis was performed. For various biological processes involved in protein modification, their Benjamini Hochberg corrected p value (as -Log10(FDR)) and enrichment factor are shown. The dashed line indicates the significance cutoff at an FDR of 0.03. B, Co-modified proteins annotated among the biological processes of protein SUMOylation or deSUMOylation were identified and their interactions based on the STRING database as shown. Larger circle size corresponds to a lower q value and darker color indicates a higher difference. C, like B, except containing co-modified proteins annotated with the biological processes of protein ubiquitylation or deubiquitylation. D, like B, but showing co-modified proteins annotated to the biological processes of protein phosphorylation and dephosphorylation. E, Co-modified proteins annotated to the more general process of protein modification that were not shown in B–D, including targets involved in protein (de)methylation (orange border) and (de)acetylation (triangles). Larger shape size corresponds to a lower q value and darker color indicates a higher difference.