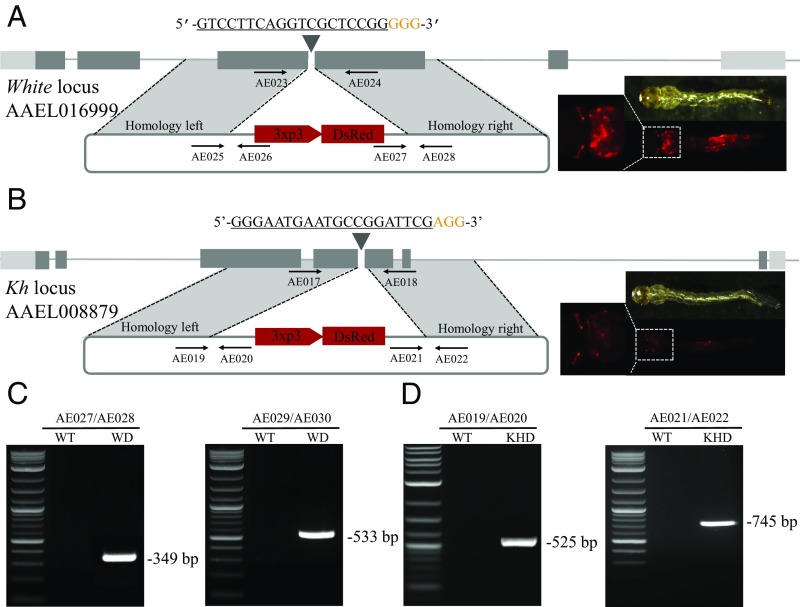
Fig. 4.
Highly efficient site-specific integration via CRISPR-mediated HDR. Schematic representations of the white locus and white-donor construct (A), and the kh locus and kh-donor construct (B). Exons are shown as boxes, coding regions are depicted in black and the 5′ and 3′ UTRs in gray. Locations and sequences of the sgRNA targets are indicated with the PAM shown in yellow. Black arrows indicate approximate positions and directions of the oligonucleotide primers used in the study. The donor plasmids (blue) express fluorescent eye marker (3xp3-DsRed) inserted between regions of homology from the white and kh locus, respectively (A and B). Gene amplification analysis confirms site-specific integration of the white-donor construct into the white locus using combinations of genomic and plasmid donor-specific primers (933Cms3/933Cms4 expected 349 bp, and 933Cms5/933Cms6 expected 533 bp) (C), and also confirms the integration of the kh-donor construct into the kh locus using combinations of genomic and plasmid donor specific primers (924ms3/924ms4 expected 525 bp, and 924ms5/924ms6 expected 745 bp) with no amplification in wild-type (D). WT represents wild-type, WD represents knockin with white-donor, KHD represents knockin with kh-donor. (Magnification: 20×.)