Fig. 5.
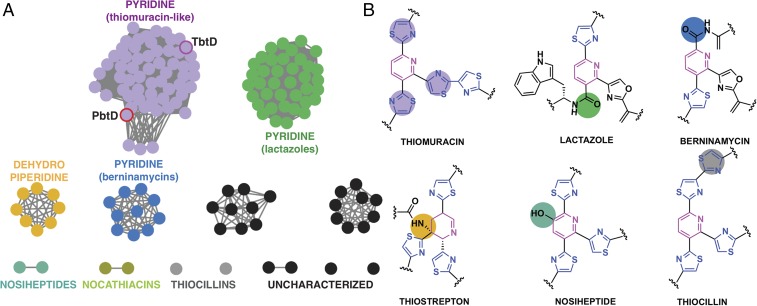
(A) Sequence similarity network of [4+2] cycloaddition proteins found across various thiopeptide biosynthetic clusters showing that the sequences cluster based on the nature of the central nitrogenous ring. Nodes represent protein sequences while edges denote sequences with at least ∼30% amino acid identity. PbtD and TbtD are highlighted. The macrocycle compositions are as follows: thiostrepton-like, 26-membered dehydropiperidines; thiocillin/nosiheptide-like, 26-membered pyridines; thiomuracin/GE2270-like, 29-membered pyridine; lactazole-like, 32-membered pyridine; berninamycin-like, 35-membered pyridine. (B) Structures of the central ring found in different thiopeptide natural products with colored circles to indicate changes in the substitution pattern. The colors correspond with the clusters shown in A. It is noted that the nocathiacin and nosiheptide [4+2] enzymes form two unique clusters, despite these thiopeptides bearing high structural homology and an identical pattern of pyridine substitution. This is likely due to limited sample size and some variation at the N terminus between sequences in these clusters.