Fig. 1.
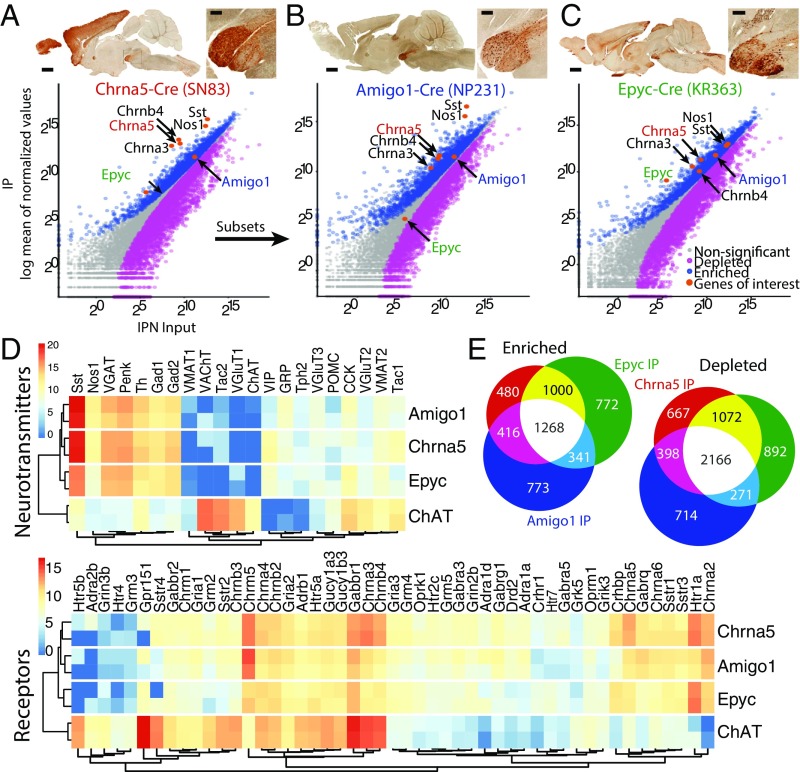
TRAP profiling reveals two subsets of α5 neuronal populations in the IPN. (A) Chrna5-Cre mice (Top) crossed to EGFP-L10a reporter mice were used for TRAP. (Bottom) Scatterplots of average TRAP IP samples (y axis) versus input samples (x axis) represent enriched (>0.6 log2 fold change, blue) or depleted (less than −0.6 log2 fold change, magenta) translated mRNAs. Highly enriched mRNAs for Amigo1 and Epyc were identified for further characterization. (B) Amigo1-Cre::EGFP-L10a mice (C) and Epyc-Cre::EGFP-L10a mice were employed for TRAP and showed enriched mRNAs for Chrna5-Chrna3-Chrnb4 as indicated in the corresponding scatterplots. (A–C) (Scale bar: low magnification 1 mm, high magnification 200 µm.) Images courtesy of GENSAT.org. (D) Expression levels (z-score transformed normalized counts) of receptors and neurotransmitters in the three IPN populations as well as in ChAT MHb neurons. (E) α5-Amigo1 and α5-Epyc cells share a large number of enriched and depleted mRNAs with Chrna5 cells.