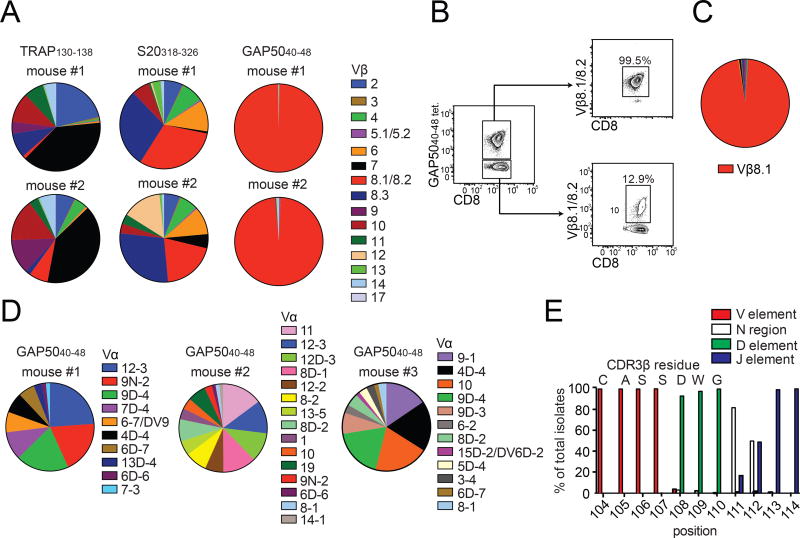
Figure 1. GAP5040–48-specific CD8+ T cells express biased TCRs.
(A, B) Mice were primed with peptide-coated DCs and boosted on day 7 with recombinant L. monocytogenes expressing the same epitope. (A) The TCR Vβ repertoire was assessed using splenocytes isolated from immunized mice by staining with the indicated tetramers and Vβ-specific antibodies. Results are shown for two mice for each antigen specificity. Data represent four independent experiments (n = 5 mice/group). Cumulative data are shown in Supplementary Figure 1B. (B) Extreme focusing of the GAP5040–48-specific repertoire does not reflect the overall frequency of Vβ8.1/8.2+ clonotypes in the CD8+ T cell compartment. Data represent three independent experiments (n = 4–5 mice/group). (C, D) Confirmation of TCR bias at the transcriptional level. Sequences were derived from GAP5040–48-specific CD8+ T cells isolated from the spleens of mice previously immunized with DC-LM-GAP5040–48 (total = 316 molecular clones). (C) Population-level analysis revealed an extreme bias towards Vβ8.1. (D) TCR Vα sequence distributions from three different mice are depicted. No obvious bias was apparent in the Vα repertoire. (E) Origin of CDR3β amino acids expressed relative to the total number of molecular Vβ8.1+ clones (n = 5 mice). Vβ: red; Dβ: green; Jβ: purple; N segments: white. (See also Figures S1–S3 and Table S1).