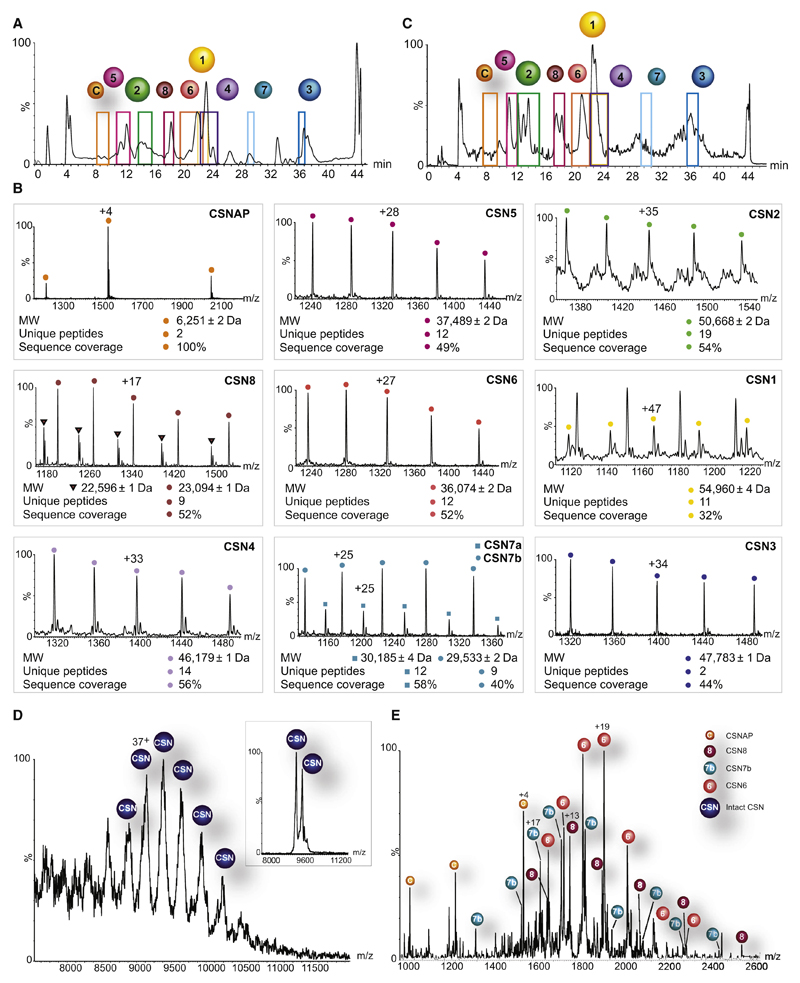
Figure 1. CSNAP physically associates with the CSN complex.
The endogenous CSN complex isolated from human erythrocytes (A) and HEK293 cells (C) was separated into its component subunits, using a monolithic column under denaturing conditions. A colored frame highlights the retention time of each eluted protein. CSNAP (labeled as C) was persistently detected alongside the eluted CSN subunits, implying its association with the CSN complex. (B) The resulting ESI-QToF mass spectra are shown. These spectra made it possible to determine the mass of the eluted protein and its associated variants; proteomics analysis was performed for sequence identification. CSNAP repeatedly co-eluted with the CSN subunits, suggesting that it associates with the complex. Indicated masses are an average of biological and technical measurements. Subunit variants are differentiated by labeling with circles and squares. (D) Nano-electrospray mass spectrum recorded under native conditions of the human CSN complex isolated from HEK293T cells. The intact CSN complex is observed between 8,500 and 10,500 m/z. The 36+ and 35+ charge states (inset) were selected for tandem MS analysis. (E) MS/MS spectrum showing the individual subunits stripped from the CSN complex. In addition to the dissociation of CSN6, CSN7b, and CSN8, we could also assign peaks corresponding in mass to CSNAP, indicating that it interacts with the CSN complex. The different species are denoted with labeled circles.