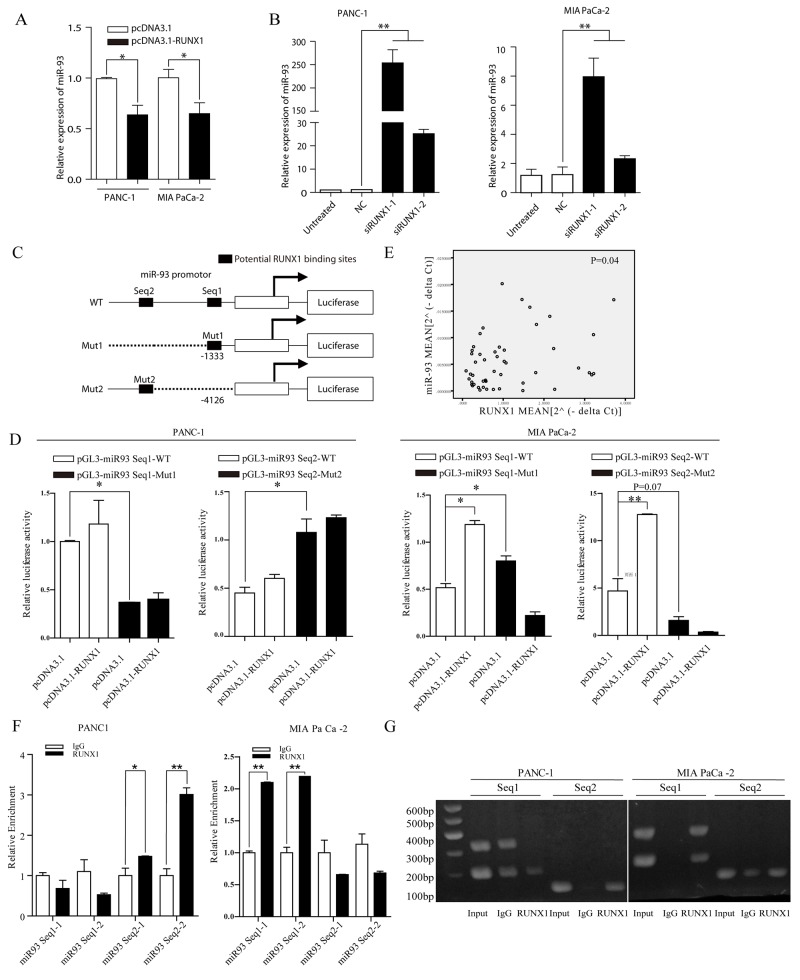
Figure 3.
(A) miR-93 expression decreased in pcDNA3. 1-RUNX1 transefected PANC-1 and MIA PaCa-2 cells, compared with pcDNA3.1 group. Data were normalized by U6. *P<0.05. (B) Relative miR-93 expression level measured by RT-qPCR. Two cells were separately transfected with NC and two RUNX1 siRNA (siRUNX1-1, siRUNX1-2). Results were normalized by U6. ** P < 0.01. (C) Schematic diagram of the pGL3 luciferase reporter constructs containing miR-93 precurser and upstream two potential RUNX1 binding sites (-1333 and -4126). The black boxes indicate the two predicted RUNX1 binding site in the promoter region of miR-93, respectively. The white box indicates miR-93 precurser. Seq1 and Seq2 refer to each sequence long enough to cover binding sites for primers use or sequence insert). (D) The luciferase reporter constructs of the truncated miR-93 promoters and corresponding mutates were introduced into PANC-1 and MIAPaCa-2 cells pretransfected with pcDNA3.1-RUNX1 or empty. Luciferase activity was measured 48 h post transfection. (E) Expression of miR-93 was positively correlated with that of RUNX1 (P=0.04) in clinical samples. (F) Fold-enrichment of RUNX1 binding at the miR-93 promoter relative to background in PANC-1 and MIA PaCa-2 cells was measured by RT-qPCR. Normalized to GAPDH, results were adjusted as n-fold compared to IgG. Data are presented as mean ± SEM. ** P<0.01. (G) Agarose gel analysis of ChIP-PCR products.