Abstract
The taxonomy of the coelomycetes has undergone dramatic changes in recent years, but remains controversial due to the high number of taxa involved, their poor morphological differentiation, the rare occurrence of the sexual morphs, and rapid loss of fertility in vitro. In the present study, we revisited the families Cucurbitariaceae and Didymellaceae (Pleosporales, Dothideomycetes), which include numerous plant pathogens, endophytic species associated with a wide host range, and saprobes. The taxonomy of two of the most relevant genera, i.e. Phoma and Pyrenochaeta, remains ambiguous after several phylogenetic studies, and needs further revision. We have studied a total of 143 strains of coelomycetes from clinical or environmental origin, by combining the LSU, ITS, tub2 and rpb2 sequences for a multi-locus analysis and a detailed morphological comparison. The resulting phylogenetic tree revealed that some fungi previously considered as members of Cucurbitariaceae represented five different families, and four of them, Neopyrenochaetaceae, Parapyrenochaetaceae, Pseudopyrenochaetaceae and Pyrenochaetopsidaceae, are proposed here as new. Furthermore, 13 new genera, 28 new species, and 20 new combinations are proposed within the Pleosporineae. Moreover, four new typifications are introduced to stabilise the taxonomy of these fungi.
Key words: Cucurbitariaceae, Didymellaceae, Multigene phylogeny, New taxa, Phoma, Pleosporineae, Pleosporales, Pyrenochaeta, Pyrenochaetopsis, Taxonomy
Taxonomic novelties: New families: Neopyrenochaetaceae Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Parapyrenochaetaceae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Pseudopyrenochaetaceae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Pyrenochaetopsidaceae Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel
New genera: Allocucurbitaria Valenzuela-Lopez, Stchigel, Guarro & Cano; Cumuliphoma Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Ectophoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Juxtiphoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Neopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neopyrenochaetopsis Valenzuela-Lopez, Cano, Guarro & Stchigel; Paracucurbitaria Valenzuela-Lopez, Stchigel, Guarro & Cano; Parapyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Pseudopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Remotididymella Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Similiphoma Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Vacuiphoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Xenopyrenochaetopsis Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano
New species: Allocucurbitaria botulispora Valenzuela-Lopez, Stchigel, Guarro & Cano; Allophoma cylindrispora Valenzuela-Lopez, Cano, Guarro & Stchigel; Cumuliphoma indica Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Cu. pneumoniae Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Didymella brunneospora Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; D. keratinophila Valenzuela-Lopez, Cano, Guarro & Stchigel; Epicoccum catenisporum Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Ep. keratinophilum Valenzuela-Lopez, Cano, Guarro & Stchigel; Ep. ovisporum Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Ep. pneumoniae Valenzuela-Lopez, Stchigel, Guarro & Cano; Neoascochyta cylindrispora Valenzuela-Lopez, Cano, Guarro & Stchigel; Neoa. tardicrescens Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Neocucurbitaria aquatica Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neocu. irregularis Valenzuela-Lopez, Cano, Guarro & Stchigel; Neopyrenochaeta fragariae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neopyrenochaetopsis hominis Valenzuela-Lopez, Cano, Guarro & Stchigel; Nothophoma variabilis Valenzuela-Lopez, Cano, Guarro & Stchigel; Paracucurbitaria italica Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Pseudopyrenochaeta terrestris Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Pyrenochaetopsis americana Valenzuela-Lopez, Cano, Guarro & Stchigel; Py. botulispora Valenzuela-Lopez, Cano, Guarro & Stchigel; Py. confluens Valenzuela-Lopez, Cano, Guarro & Stchigel; Py. globosa Valenzuela-Lopez, Cano, Guarro & Stchigel; Py. paucisetosa Valenzuela-Lopez, Cano, Guarro & Stchigel; Py. setosissima Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Py. uberiformis Valenzuela-Lopez, Cano, Guarro & Stchigel; Remotididymella anthropophila Valenzuela-Lopez, Cano, Guarro & Stchigel; Vacuiphoma oculihominis Valenzuela-Lopez, Stchigel, Guarro & Cano
New combinations: Cumuliphoma omnivirens (Aveskamp et al.) Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Ectophoma multirostrata (P.N. Mathur et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Ec. pomi (Horne) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Epicoccum proteae (Crous) Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano; Juxtiphoma eupyrena (Sacc.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Neocucurbitaria cava (Schulzer) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neocu. hakeae (Crous) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neocu. keratinophila (Verkley et al.) Valenzuela-Lopez, Stchigel, Guarro & Cano; Neopyrenochaeta acicola (Moug. & Lév.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neopy. inflorescentiae (Crous et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Neopy. telephoni (Rohit Sharma et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Paracucurbitaria corni (Bat. & A.F. Vital) Valenzuela-Lopez, Stchigel, Guarro & Cano; Parapyrenochaeta acaciae (Crous et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Parapy. protearum (Crous) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Pseudopyrenochaeta lycopersici (R.W. Schneid. & Gerlach) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano; Remotididymella destructiva (Plowr.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Similiphoma crystallifera (Gruyter et al.) Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Vacuiphoma bulgarica (Aveskamp et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel; Xenodidymella saxea (Aveskamp et al.) Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel; Xenopyrenochaetopsis pratorum (P.R. Johnst. & Boerema) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano
Introduction
The Pleosporales is the largest order of the class Dothideomycetes (phylum Ascomycota), encompassing more than 4 700 species distributed over 332 genera, and 53 families (Kirk et al., 2008, Zhang et al., 2009, Zhang et al., 2012, Ariyawansa et al., 2013, Hyde et al., 2013, Amaradasa et al., 2014, Trakunyingcharoen et al., 2014, Wijayawardene et al., 2014, Crous et al., 2015a, Sharma et al., 2015, Tanaka et al., 2015, Jaklitsch et al., 2016, Jaklitsch and Voglmayr, 2016, Wanasinghe et al., 2016, Crous and Groenewald, 2017, Hashimoto et al., 2017, Hernández-Restrepo et al., 2017). These fungi are characterised by the production of pseudothecial ascomata (mostly globose and usually papillate) consisting of a peridial wall composed by several layers of cells, within which the fissitunicate (bitunicate) asci are produced amidst a persistent hamathecium (the vegetative structures inside an ascoma) (Jaklitsch and Voglmayr, 2016, Jaklitsch et al., 2017, Zhang et al., 2009, Zhang et al., 2012) and ascospores, which are mostly septate but variable in shape and pigmentation. The asexual morphs of the Pleosporales are characterised by conidia produced within discrete sporocarps (conidiomata), and sometimes conidia are generated on conidiophores produced on mycelium. Phoma and its relatives are the most common pleosporalean asexual morphs and are characterised by the presence of pycnidia (globose to pyriform conidiomata from which the conidia arise throughout an apical opening) (De Gruyter et al., 2009, De Gruyter et al., 2010, Aveskamp et al., 2010, Chen et al., 2015). Pleosporales are mainly saprobic on plant debris, epiphytic, endophytic or parasitic of living plants, fungi and insects, or mycobionts in lichens (Kruys et al., 2006, Aveskamp et al., 2008, Aveskamp et al., 2010, De Gruyter et al., 2009, Zhang et al., 2009, Zhang et al., 2012, Lawrey et al., 2012, Kocakaya et al., 2015). These fungi can also infect humans (Punithalingam, 1979, Ahmed et al., 2014, Ahmed et al., 2015, Ahmed et al., 2017, Borman et al., 2016, Valenzuela-Lopez et al., 2016).
Modern phylogenetic studies support the division of the Pleosporales into the suborders Pleosporineae and Massarineae (Zhang et al., 2009, Zhang et al., 2012, Hyde et al., 2013, Tanaka et al., 2015). The former includes nine families, i.e. Coniothyriaceae, Cucurbitariaceae, Didymellaceae, Dothidotthiaceae, Halojulellaceae, Leptosphaeriaceae, Neophaeosphaeriaceae, Phaeosphaeriaceae, Pleosporaceae and Shiraiaceae (Zhang et al., 2012, De Gruyter et al., 2013, Ariyawansa et al., 2013, Ariyawansa et al., 2015b, Liu et al., 2013), which encompass plant pathogens of economic importance including the well-known genera such as Alternaria, Ascochyta, Bipolaris, Didymella and Leptosphaeria (Zhang et al., 2012, Ariyawansa et al., 2013, De Gruyter et al., 2013, Liu et al., 2013, Woudenberg et al., 2013). Recently, Tanaka et al. (2015) revised the suborder Massarineae and accepted 12 families; however, more studies are needed for a better understanding of their phylogenetic relationships. Numerous species of Pleosporales are relatively common in clinical samples, most of which belong to the families Cucurbitariaceae and Didymellaceae (Valenzuela-Lopez et al. 2016). Cucurbitariaceae is still a poorly known family, which was erected by Winter (1885) with Cucurbitaria as the type genus, and characterised by ostiolate ascomata aggregated on a basal pseudostromatic structure, hamathecium composed of wide persistent filaments, fissitunicate, cylindrical to cylindrical-clavate asci and dark, phragmosporous or muriform ascospores. In the last revision of Cucurbitariaceae, four sexual genera (Cucurbitaria, Curreya, Rhytidiella and Syncarpella) and two asexual genera (Pyrenochaeta and Pyrenochaetopsis) were accepted (Doilom et al. 2013). The latter two genera are characterised by phoma-like, setose pycnidia, and hyaline, aseptate conidia (De Gruyter et al., 2010, De Gruyter et al., 2013). Recently, Jaklitsch & Voglmayr (2017) demonstrated that some species of Cucurbitaria, such as C. obducens, C. piceae (both producing muriform ascospores) and C. rhododendri (with phragmospores), belong to three different genera of Melanommataceae. Wanasinghe et al. (2017b) proposed Neocucurbitaria, characterised by solitary ascomata, the presence of periphyses and muriform ascospores, as a new genus of Cucurbitariaceae. However, the current members of this family need to be re-evaluated, including their asexual morphs.
The family Didymellaceae also includes economically important plant pathogens, such as the causal agents of blackleg and ascochyta blight (Rouxel and Balesdent, 2005, McDonald and Peck, 2009, Salam et al., 2011, De Gruyter et al., 2013), but also diverse endophytic, fungicolous and lichenicolous taxa belong to this fungal group (Aveskamp et al. 2010), whereas a few members are known as pathogens of humans (de Hoog et al. 2011). This family was established by de Gruyter et al. (2009) and embraces the species traditionally classified in the genera Ascochyta, Didymella and Phoma. However, Phoma is one of the largest and most polyphyletic fungal genera (with more than 3 000 names recorded) with species occurring in more than 25 families (http://www.indexfungorum.org).
Zhang et al. (2009), included Didymellaceae in their study and accepted the sexual genera Didymella, Leptosphaerulina, Macroventuria, Monascostroma and Platychora. In general, these genera are characterised by dark pseudothecial ascomata, filamentous pseudoparaphyses, 8-spored, fissitunicate, clavate to saccate asci, and hyaline, 1-septate, fusiform to biconical ascospores; with the only exception being Leptosphaerulina, which has hyaline to brown, ellipsoid, cylindrical or oblong, phragmosporous or muriformly septate ascospores, which also lack pseudoparaphyses. Several studies have tried to resolve the taxonomy of the asexual morphs of the Didymellaceae, especially Phoma and its relatives, with more or less success. Subsequently, de Gruyter et al. (2010) transferred several species of Phoma to Pyrenochaetopsis (Cucurbitariaceae), Neosetophoma and Setophoma (Phaeosphaeriaceae), and resurrected the genus Paraphoma (Phaeosphaeriaceae). The study by Aveskamp et al. (2010), based on the sequences of four loci, revealed that the subdivision of Phoma in sections (Boerema et al. 2004) was phylogenetically inconsistent, and they thus proposed Boeremia to accommodate species morphologically close to Phoma exigua, while species of Phoma section Sclerophomella were transferred to Epicoccum and Peyronellaea. Furthermore, de Gruyter et al. (2013) transferred some species of Phoma sections Plenodomus and Heterospora to the Leptosphaeriaceae and some from Phoma section Pilosa and Ascochyta to Pleosporaceae. Recently, Chen et al. (2015) proposed nine genera (Allophoma, Calophoma, Heterophoma, Neoascochyta, Neodidymelliopsis, Nothophoma, Paraboeremia, Phomatodes and Xenodidymella) in Didymellaceae, transferred Microsphaeropsis (Didymellaceae) to the family Microsphaeropsidaceae, and restricted Phoma to Phoma herbarum (Chen et al. 2017). Other authors have added the genera Briansuttonomyces, Didymellocamarosporium, Heracleicola, Neodidymella, Neomicrosphaeropsis and Pseudoascochyta to Didymellaceae (Ariyawansa et al., 2015a, Crous and Groenewald, 2016, Crous et al., 2016a, Thambugala et al., 2016, Wijayawardene et al., 2016). However, the genera Didymellocamarosporium, Heracleicola and Neodidymella were studied by Chen et al. (2017) and revealed as probable synonyms of older genera within Didymellaceae.
To resolve the taxonomy of the Cucurbitariaceae and the Didymellaceae we have tried to delineate the phylogenetic relationships within these families performing a multi-locus analysis including ex-type and reference strains of most of the phoma-like and pyrenochaeta-like taxa available in the culture collection of Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands; formerly CBS-KNAW), and numerous isolates of clinical origin from the USA.
Materials and methods
Isolates and reference fungal strains
This study comprised 70 clinical isolates previously identified as belonging to the Pleosporales (Valenzuela-Lopez et al. 2016), provided by the Fungus Testing Laboratory of the University of Texas Health Science Center at San Antonio (UTHSC; San Antonio, Texas, USA), two environmental strains from Spain (CBS 141688) and New Zealand (CBS 141689) respectively, and 71 reference and ex-type strains belonging to the Cucurbitariaceae and Didymellaceae provided by the CBS culture collection (Table 1).
Table 1.
Isolates used in this study and their GenBank accession numbers. Numbers of new taxa, combinations and sequences generated are indicated in bold.
| Species | Old name | CBS strain1 no. | Other strain1 no. | Status2 | Host, substrate | Country | GenBank accession numbers3 |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| LSU | ITS | TUB | RPB2 | |||||||
| Allocucurbitaria botulispora | Pyrenochaeta sp. | CBS 142452 | UTHSC:DI16-273; FMR 13764 | T | Human superficial tissue | USA | LN907416 | LT592932 | LT593001 | LT593070 |
| Allophoma cylindrispora | Phoma sp. | CBS 142453 | UTHSC:DI16-233; FMR 13723 | T | Human superficial tissue | USA | LN907376 | LT592920 | LT592989 | LT593058 |
| A.labilis | CBS 124.93 | Solanum lycopersicum | The Netherlands | GU238091 | GU237765 | GU237619 | KT389552 | |||
| A.minor | CBS 325.82 | FMR 14905 | T | Syzygium aromaticum | Indonesia | GU238107 | GU237831 | GU237632 | KT389553 | |
| A.nicaraguensis | CBS 506.91 | FMR 14904 | T | Coffea arabica | Nicaragua | GU238058 | GU237876 | GU237596 | KT389551 | |
| A.oligotrophica | Phoma costarricencis | CBS 497.91 | FMR 14902 | Coffea arabica | Unknown | GU238059 | GU237870 | GU237597 | LT623247 | |
| CGMCC 3.18114 | T | Air sample | China | KY742194 | KY742040 | KY742282 | KY742128 | |||
| A.piperis | CBS 268.93 | T | Peperomia pereskiifolia | Netherlands | GU238129 | GU237816 | GU237644 | KT389554 | ||
| A.tropica | CBS 436.75 | FMR 14903 | T | Saintpaulia ionantha | Germany | GU238149 | GU237864 | GU237663 | KT389556 | |
| A.zantedeschiae | CBS 131.93 | Calla sp. | The Netherlands | GU238159 | FJ427084 | FJ427188 | KT389557 | |||
| CBS 229.32 | Cicer arietinum | Romania | KT389690 | KT389473 | KT389558 | KT389767 | ||||
| Alternariaster bidentis | CBS 134021 | CPC 19479 | T | Bidens sulphurea | Brazil | KC609341 | KC609333 | – | KC609347 | |
| A.helianthi | CBS 327.69 | IFO 9089 | Helianthus annuus | Unknown | KC584369 | KC609335 | – | KC584494 | ||
| Ascochyta herbicola | CBS 629.97 | Water | USA | GU238083 | GU237898 | GU237614 | KP330421 | |||
| A.pisi | CBS 126.54 | AFTOL-ID 1583 | Pisum sativum | The Netherlands | DQ678070 | GU237772 | GU237531 | DQ677967 | ||
| A.rabiei | CBS 206.30 | Unknown | Unknown | KT389695 | KT389478 | KT389772 | KT389559 | |||
| A.versabilis | CBS 876.97 | Silene sp. | The Netherlands | GU238152 | GU237909 | GU237664 | KT389561 | |||
| A.viciae | CBS 451.68 | Vicia sepium | The Netherlands | KT389701 | KT389484 | KT389778 | KT389562 | |||
| Boeremia exigua | CBS 118.38 | Cheiranthus cheiri | Denmark | KT389706 | KT389489 | KT389783 | KT389582 | |||
| CBS 119.38 | Nicotiana tabacum | Unknown | KT389707 | KT389490 | KT389784 | KT389583 | ||||
| B.lycopersici | CBS 378.67 | Solanum lycopersicum | The Netherlands | GU237950 | GU237848 | GU237512 | KT389580 | |||
| Briansuttonomyces eucalypti | CBS 114879 | CPC 362 | T | Eucalyptus sp. | South Africa | KU728519 | KU728479 | KU728595 | – | |
| CBS 114887 | CPC 363 | Eucalyptus sp. | South Africa | KU728520 | KU728480 | KU728596 | – | |||
| Calophoma aquilegiicola | CBS 107.96 | Aconitum pyramidale | The Netherlands | GU238041 | GU237735 | GU237581 | KT389586 | |||
| CBS 108.96 | Aquilegia sp. | The Netherlands | GU238042 | GU237736 | GU237582 | – | ||||
| C.clematidina | CBS 102.66 | Clematis sp. | UK | FJ515630 | FJ426988 | FJ427099 | KT389587 | |||
| CBS 108.79 | T | Clematis sp. | The Netherlands | FJ515632 | FJ426989 | FJ427100 | KT389588 | |||
| C.clematidis-rectae | CBS 507.63 | T | Clematis sp. | The Netherlands | FJ515647 | FJ515606 | FJ515624 | KT389589 | ||
| C.rosae | CGMCC 3.18347 | T | Rosa sp. | China | KY742203 | KY742049 | KY742291 | KY742135 | ||
| LC 8119 | Rosa sp. | China | KY742204 | KY742050 | KY742292 | KY742136 | ||||
| Camarosporidiella aborescentis | Camarosporium aborescentis | MFLUCC 14-0604 | T | Colutea arborescens | Russia | KP711378 | KP711377 | – | – | |
| Camarosporium arezzoensis | MFLUCC 14-0238 | T | Cytisus sp. | Italy | KP120927 | KP120926 | – | – | ||
| C.aureum | Camarosporium aureum | MFLUCC 14-0620 | T | Cotinus coggygria | Russia | KP744478 | KP744436 | – | – | |
| C.clematidis | Camarosporium clematidis | MFLUCC 13-0336 | T | Clematis vitalba | Italy | KJ562188 | KJ562213 | – | – | |
| C.elongata | Cucurbitaria elongata | MFLUCC 14-0260 | Cytisus scoparius | Italy | KJ724249 | – | – | – | ||
| Cucurbitaria elongata | CBS 171.55 | AFTOL-ID 1568 | Cytisus sessilifolius | France | DQ678061 | – | – | DQ677957 | ||
| C.robiniicola | Camarosporium robiniicola | MFLUCC 13-0527 | Robinia pseudacacia | Italy | KJ589412 | KJ562214 | – | – | ||
| C.spartii | Camarosporium spartii | MFLUCC 13-0548 | Cytisus sp. | Italy | KJ589413 | KJ562215 | – | – | ||
| Camarosporium quaternatum | CBS 142617 | CPC 23216 | Daphne mezereum | Germany | KY929170 | KY929135 | – | – | ||
| CBS 142616 | CPC 31081 | T | Lycium barbarum | Hungary | KY929136 | KY929171 | – | – | ||
| CPC 31518 | Lycium barbarum | Hungary | KY929172 | KY929137 | – | – | ||||
| Camarosporomyces flavigenus | CBS 314.80 | T | Water | Romania | GU238076 | KY929138 | – | – | ||
| Coniothyrium palmarum | CBS 758.73 | CMW 5283 | Phoenix dactylifera | Israel | JX681085 | – | – | – | ||
| CBS 400.71 | Chamaerops humilis | Italy | EU754153 | AY720708 | KT389792 | KT389592 | ||||
| C.telephii | CBS 188.71 | Air sample | Finland | GQ387599 | JF740188 | KT389793 | KT389593 | |||
| CBS 856.97 | Mineral wool | Finland | GQ387600 | JF740189 | ||||||
| Cucurbitaria berberidis | MFLUCC 11-0387 | Berberis vulgaris | Austria | KC506796 | – | – | – | |||
| CBS 130007 | FMR 15751; MFLUCC 11-0384; CB1 | T | Berberis vulgaris | Austria | KC506793 | LT717673 | LT717676 | LT854936 | ||
| Cumuliphoma indica | Phoma omnivirens | CBS 654.77 | FMR 15341 | T | Unknown | India | GU238122 | FJ427043 | FJ427153 | LT623261 |
| Phoma omnivirens | CBS 991.95 | FMR 15331 | Soil | Papua New Guinea | GU238121 | FJ427044 | FJ427154 | LT623262 | ||
| C.omnivirens | Phoma omnivirens | CBS 341.86 | FMR 14915 | T | Phaseolus vulgaris | Belgium | LT623214 | FJ427042 | FJ427152 | LT623260 |
| C.pneumoniae | Phoma sp. | CBS 142454 | UTHSC:DI16-249; FMR 13739 | T | Human respiratory tract | USA | LN907392 | LT592925 | LT592994 | LT593063 |
| Cucurbidothis pityophila | CBS 149.32 | FMR 15744 | Unknown | The Netherlands | JX681087 | GQ203756 | LT854934 | LT854935 | ||
| Didymella aeria | LC 8120 | Air sample | China | KY742206 | KY742052 | KY742294 | KY742138 | |||
| CGMCC 3.18353 | T | Air sample | China | KY742205 | KY742051 | KY742137 | KY742293 | |||
| D.aliena | CBS 379.93 | Berberis sp. | The Netherlands | GU238037 | GU237851 | GU237578 | KP330416 | |||
| D.americana | CBS 185.85 | Zea mays | USA | GU237990 | FJ426972 | FJ427088 | KT389594 | |||
| D.anserina | CBS 253.80 | Unknown | Germany | KT389715 | KT389498 | KT389795 | KT389595 | |||
| Peyronellaea sp. | UTHSC:DI16-255; FMR 13745 | Human respiratory tract | USA | LN907398 | LT592926 | LT592995 | LT593064 | |||
| D.aquatica | CGMCC 3.18349 | T | Water | China | KY742209 | KY742055 | KY742297 | KY742140 | ||
| LC 5555 | Water | China | KY742210 | KY742056 | KY742298 | KY742141 | ||||
| D.arachidicola | CBS 333.75 | T | Arachis hypogaea | South Africa | GU237996 | GU237833 | GU237554 | KT389598 | ||
| D.aurea | CBS 269.93 | T | Medicago polymorpha | New Zealand | GU237999 | GU237818 | GU237557 | KT389599 | ||
| D.bellidis | CBS 714.85 | Bellis perennis | The Netherlands | GU238046 | GU237904 | GU237586 | KP330417 | |||
| D.boeremae | CBS 109942 | T | Medicago littoralis cv. Harbinger | Australia | GU238048 | FJ426982 | FJ427097 | KT389600 | ||
| D.brunneospora | Didymella sp. | CBS 115.58 | FMR 15745 | T | Chrysanthemum roseum | Germany | KT389723 | KT389505 | KT389802 | KT389625 |
| D.chenopodii | CBS 128.93 | Chenopodium quinoa cv. Sajana | Peru | GU238055 | GU237775 | GU237591 | KT389602 | |||
| D.chloroguttulata | CGMCC 3.18351 | T | Air sample | China | KY742211 | KY742057 | KY742299 | KY742142 | ||
| LC 8122 | Air sample | China | KY742212 | KY742058 | KY742300 | KY742143 | ||||
| D.coffeae-arabicae | CBS 123380 | PD 84/1013 | T | Coffea arabica | Ethiopia | GU238005 | FJ426993 | FJ427104 | KT389603 | |
| D.curtisii | PD 92/1460 | Sprekelia sp. | The Netherlands | GU238012 | FJ427041 | FJ427151 | KT389604 | |||
| D.ellipsoidea | CGMCC 3.18350 | T | Air sample | China | KY742214 | KY742060 | KY742302 | KY742145 | ||
| LC 8123 | Air sample | China | KY742215 | KY742061 | KY742303 | KY742146 | ||||
| D.eucalyptica | CBS 377.91 | Eucalyptus sp. | Australia | GU238007 | GU237846 | GU237562 | KT389605 | |||
| D.exigua | CBS 183.55 | T | Rumex arifolius | France | EU754155 | GU237794 | GU237525 | EU874850 | ||
| D.gardeniae | CBS 626.68 | IMI 108771; FMR 14901 | T | Gardenia jasminoides | India | GQ387595 | FJ427003 | FJ427114 | KT389606 | |
| Peyronellaea sp. | UTHSC:DI16-211; FMR 13701 | Human superficial tissue | USA | LN907354 | LT592908 | LT592977 | LT593046 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-226; FMR 13716 | Human superficial tissue | USA | LN907369 | LT592913 | LT592982 | LT593051 | |||
| Peyronellaea sp. | UTHSC:DI16-274; FMR 13765 | Human superficial tissue | USA | LN907417 | LT592933 | LT593002 | LT593071 | |||
| Peyronellaea sp. | UTHSC:DI16-295; FMR 13788 | Human superficial tissue | USA | LN907438 | LT592944 | LT593013 | LT593083 | |||
| D.glomerata | CBS 528.66 | T | Chrysanthemum sp. | The Netherlands | JX681105 | FJ427013 | FJ427124 | GU371781 | ||
| Peyronellaea glomerata | UTHSC:DI16-205; FMR 13695 | Human superficial tissue | USA | LN907348 | LT592905 | LT592974 | LT593043 | |||
| D.heteroderae | CBS 109.92 | PD 73/1405 | T | Undefined food material | The Netherlands | GU238002 | FJ426983 | FJ427098 | KT389601 | |
| Peyronellaea calorpreferens | UTHSC:DI16-190; FMR 13680 | Human superficial tissue | USA | LN907333 | LT592896 | LT592965 | LT593034 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-224; FMR 13714 | Human superficial tissue | USA | LN907367 | LT592911 | LT592980 | LT593049 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-227; FMR 13717 | Human superficial tissue | USA | LN907370 | LT592914 | LT592983 | LT593052 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-231; FMR 13721 | Human superficial tissue | USA | LN907374 | LT592918 | LT592987 | LT593056 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-232; FMR 13722 | Human deep tissue/fluids | USA | LN907375 | LT592919 | LT592988 | LT593057 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-234; FMR 13724 | Human superficial tissue | USA | LN907377 | LT592921 | LT592990 | LT593059 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-235; FMR 13725 | Human superficial tissue | USA | LN907378 | LT592922 | LT592991 | LT593060 | |||
| Peyronellaea calorpreferens | UTHSC:DI16-305; FMR 13798 | Human respiratory tract | USA | LN907448 | LT592951 | LT593020 | LT593090 | |||
| D.ilicicola | CGMCC 3.18355 | T | Ilex chinensis | Italy | KY742219 | KY742065 | KY742307 | KY742150 | ||
| LC 8127 | Ilex chinensis | Italy | KY742220 | KY742066 | KY742308 | KY742151 | ||||
| D.infuscatispora | CGMCC 3.18356 | T | Chrysanthemum indicum | China | KY742221 | KY742067 | KY742309 | KY742152 | ||
| LC 8129 | Chrysanthemum indicum | China | KY742222 | KY742068 | KY742310 | – | ||||
| D.keratinophila | Peyronellaea sp. | CBS 143032 | UTHSC:DI16-200; FMR 13690 | T | Human superficial tissue | USA | LN907343 | LT592901 | LT592970 | LT593039 |
| Peyronellaea sp. | UTHSC:DI16-228; FMR 13718 | Human superficial tissue | USA | LN907371 | LT592915 | LT592984 | LT593053 | |||
| Phoma sp. | UTHSC:DI16-282; FMR 13774 | Human superficial tissue | USA | LN907425 | LT592938 | LT593007 | LT593077 | |||
| D.lethalis | CBS 103.25 | Unknown | Unknown | GU238010 | GU237729 | GU237564 | KT389607 | |||
| D.macrophylla | CGMCC 3.18357 | T | Hydrangea macrophylla | Italy | KY742224 | KY742070 | KY742312 | KY742154 | ||
| LC 8132 | Hydrangea macrophylla | Italy | KY742225 | KY742071 | KY742313 | KY742155 | ||||
| D.macrostoma | CBS 223.69 | Acer pseudoplatanus | Switzerland | GU238096 | GU237801 | GU237623 | KT389608 | |||
| D.maydis | CBS 588.69 | T | Zea mays | USA | EU754192 | FJ427086 | FJ427190 | GU371782 | ||
| D.microchlamydospora | CBS 105.95 | T | Eucalyptus sp. | UK | GU238104 | FJ427028 | FJ427138 | KP330424 | ||
| Phoma sp. | UTHSC:DI16-199; FMR 13689 | Human superficial tissue | USA | LN907342 | LT592900 | LT592969 | LT593038 | |||
| Peyronellaea sp. | UTHSC:DI16-365; FMR 13858 | Human superficial tissue | USA | LN907508 | LT592964 | LT593033 | LT593103 | |||
| D.molleriana | CBS 229.79 | Digitalis purpurea | New Zealand | GU238067 | GU237802 | GU237605 | KP330418 | |||
| D.musae | Phoma sp. | UTHSC:DI16-230; FMR 13720 | Human superficial tissue | USA | LN907373 | LT592917 | LT592986 | LT593055 | ||
| CBS 463.69 | FMR 15339 | Mangifera indica | India | GU238011 | FJ427026 | FJ427136 | LT623248 | |||
| D.negriana | CBS 358.71 | Vitis vinifera | Germany | GU238116 | GU237838 | GU237635 | KT389610 | |||
| ICMP 10845; LC 5249 | Vitis vinifera | former Yugoslavia | KY742227 | KY742073 | KY742315 | – | ||||
| D.nigricans | CBS 444.81 | PD 77/919 | Actinidea chinensis | New Zealand | GU238001 | GU237915 | GU237559 | KT389611 | ||
| D.ocimicola | CGMCC 3.18358 | T | Ocimum sp. | China | KY742232 | KY742078 | KY742320 | – | ||
| LC 8138 | Ocimum sp. | China | KY742233 | KY742079 | KY742321 | – | ||||
| D.pedeiae | CBS 124517 | T | Schefflera elegantissima | The Netherlands | GU238127 | GU237770 | GU237642 | KT389612 | ||
| D.pinodella | CBS 531.66 | Trifolium pretense | USA | GU238017 | FJ427052 | FJ427162 | KT389613 | |||
| D.pinodes | CBS 525.77 | T | Pisum sativum | Belgium | GU238023 | GU237883 | GU237572 | KT389614 | ||
| CBS 374.84 | FMR 15345 | Pisum sativum | The Netherlands | EU754135 | JF810523 | LT623229 | LT623249 | |||
| D.pomorum | CBS 285.76 | T | Heracleum dissectum | Russia | GU238025 | FJ427053 | FJ427163 | KT389615 | ||
| D.protuberans | CBS 381.96 | FMR 14899 | T | Lycium halifolium | The Netherlands | GU238029 | GU237853 | GU237574 | KT389620 | |
| Peyronellaea sp. | UTHSC:DI16-302; FMR 13795 | Environmental | USA | LN907445 | LT592949 | LT593018 | LT593088 | |||
| D.pteridis | CBS 379.96 | FMR 15750 | Pteris sp. | The Netherlands | KT389722 | KT389504 | KT389801 | KT389624 | ||
| D.rhei | CBS 109177 | Rheum rhaponticum | New Zealand | GU238139 | GU237743 | GU237653 | KP330428 | |||
| D.rumicicola | Didymella acetosellae | CBS 179.97 | Rumex hydrolapathum | The Netherlands | GU238034 | GU237793 | GU237575 | KP330415 | ||
| CBS 683.79 | T | Rumex obtusifolius | New Zealand | KT389721 | KT389503 | KT389800 | KT389622 | |||
| D.sancta | CBS 281.83 | T | Ailanthus altissima | South Africa | GU238030 | FJ427063 | FJ427170 | KT389623 | ||
| CBS 644.97 | FMR 15351 | Opuntia ficus-indica | Argentina | LT623211 | FJ427064 | FJ427171 | LT623250 | |||
| D.segeticola | CGMCC 3.17489 | T | Cirsium segetum | China | KP330455 | KP330443 | KP330399 | KP330414 | ||
| CGMCC 3.17498 | Cirsium segetum | China | KP330454 | KP330442 | KP330398 | KP330413 | ||||
| D.sinensis | LC 8142 | Dendrobium officinale | China | KY742241 | KY742087 | KY742329 | KY742166 | |||
| LC 8143 | Dendrobium officinale | China | KY742242 | KY742088 | KY742330 | KY742167 | ||||
| Didymella sp. | Didymella segeticola | LC 8141 | Camellia sasanqua | Japan | KY742238 | KY742084 | KY742326 | KY742164 | ||
| D.subglomerata | CBS 110.92 | Triticum sp. | USA | GU238032 | FJ427080 | FJ427186 | KT389626 | |||
| D.suiyangensis | CGMCC 3.18352 | T | Air sample | China | KY742243 | KY742089 | KY742330 | KY742168 | ||
| LC 8144 | Air sample | China | KY742244 | KY742090 | KY742332 | KY742169 | ||||
| D.viburnicola | CBS 523.73 | Viburnum cassioides | The Netherlands | GU238155 | GU237879 | GU237667 | KP330430 | |||
| Dothidotthia aspera | CBS 119688 | CPC 12932 | Acer negundo | USA | EU673275 | – | – | – | ||
| D.symphoricarpi | CBS 119687 | CPC 12929 | T | Symphoricarpos rotundifolius | USA | EU673273 | – | – | – | |
| Ectophoma multirostrata | Phoma multirostrata | CBS 110.79 | FMR 15342 | Cucumis sativus | The Netherlands | GU238110 | FJ427030 | FJ427140 | LT623264 | |
| Phoma multirostrata | CBS 274.60 | FMR 15335 | T | Soil | Maharashtra | GU238111 | FJ427031 | FJ427141 | LT623265 | |
| Phoma multirostrata | CBS 368.65 | FMR 15336 | Unknown | India | GU238112 | FJ427033 | FJ427143 | LT623266 | ||
| E.pomi | Phoma pereupyrena | CBS 267.92 | FMR 15346 | T | Coffea arabica | India | GU238128 | GU237814 | GU237643 | LT623263 |
| Epicoccum brasiliense | CBS 120105 | FMR 14907 | T | Amaranthus sp. | Brazil | GU238049 | GU237760 | GU237588 | KT389627 | |
| E.camelliae | CGMCC 3.18343 | T | Camellia sinensis | China | KY742245 | KY742091 | KY742333 | KY742170 | ||
| Epicoccum sorghinum | UTHSC:DI16-201; FMR 13691 | Human respiratory tract | USA | LN907344 | LT592902 | LT592971 | LT593040 | |||
| Epicoccum sorghinum | UTHSC:DI16-202; FMR 13692 | Human superficial tissue | USA | LN907345 | LT592903 | LT592972 | LT593041 | |||
| Epicoccum sorghinum | UTHSC:DI16-206; FMR 13696 | Human superficial tissue | USA | LN907349 | LT592906 | LT592975 | LT593044 | |||
| Epicoccum sorghinum | UTHSC:DI16-280; FMR 13772 | Human superficial tissue | USA | LN907423 | LT592937 | LT593006 | LT593076 | |||
| Epicoccum sorghinum | UTHSC:DI16-338; FMR 13831 | Human superficial tissue | USA | LN907481 | LT592959 | LT593028 | LT593098 | |||
| Epicoccum sorghinum | UTHSC:DI16-345; FMR 13838 | Human subcutaneous tissue | USA | LN907488 | LT592961 | LT593030 | LT593100 | |||
| LC 4862 | Camellia sinensis | China | KY742246 | KY742092 | KY742334 | KY742171 | ||||
| E.catenisporum | Epicoccum sorghinum | CBS 181.80 | FMR 14911 | T | Oryza sativa | Guinea-Bissau | LT623213 | FJ427069 | FJ427175 | LT623253 |
| E.dendrobii | CGMCC 3.18359 | T | Dendrobium fimbriatum | China | KY742247 | KY742093 | KY742335 | – | ||
| LC 8146 | Dendrobium fimbriatum | China | KY742248 | KY74209 | KY742336 | - | ||||
| E.draconis | CBS 186.83 | FMR 14908 | Dracaena sp. | Rwanda | GU238070 | GU237795 | GU237607 | KT389628 | ||
| E.duchesneae | LC 8147 | Duchesnea indica | China | KY742250 | KY742096 | KY742338 | – | |||
| CGMCC 3.18345 | T | Duchesnea indica | China | KY742249 | KY742095 | KY742337 | – | |||
| E.henningsii | CBS 104.80 | Acacia mearnsii | Kenya | GU238081 | GU237731 | GU237612 | KT389629 | |||
| E.hordei | CGMCC 3.18360 | T | Hordeum vulgare | Australia | KY742251 | KY742097 | KY742339 | – | ||
| LC 8149 | Hordeum vulgare | Australia | KY742252 | KY742098 | KY742340 | – | ||||
| E.huancayense | CBS 105.80 | T | Solanum sp. | Peru | GU238084 | GU237732 | GU237615 | KT389630 | ||
| E.italicum | CGMCC 3.18361 | T | Acca sellowiana | Italy | KY742253 | KY742099 | KY742341 | KY742172 | ||
| LC 8151 | Acca sellowiana | Italy | KY74225 | KY742100 | KY742342 | KY742173 | ||||
| E.keratinophilum | Phoma sp. | UTHSC:DI16-244; FMR 13734 | Human superficial tissue | USA | LN907387 | LT592924 | LT592993 | LT593062 | ||
| Phoma sp. | UTHSC:DI16-258; FMR 13748 | Human respiratory tract | USA | LN907401 | LT592928 | LT592997 | LT593066 | |||
| Phoma sp. | CBS 142455 | UTHSC:DI16-271; FMR 13762 | T | Human superficial tissue | USA | LN907414 | LT592930 | LT592999 | LT593068 | |
| Phoma sp. | UTHSC:DI16-272; FMR 13763 | Human superficial tissue | USA | LN907415 | LT592931 | LT593000 | LT593069 | |||
| Phoma sp. | UTHSC:DI16-299; FMR 13792 | Human deep tissue/fluids | USA | LN907442 | LT592947 | LT593016 | LT593086 | |||
| E.latusicollum | Epicoccum sorghinum | UTHSC:DI16-197; FMR 13687 | Human superficial tissue | USA | LN907340 | LT592898 | LT592967 | LT593036 | ||
| CGMCC 3.18346 | T | Sorghum bicolor | China | KY742255 | KY742101 | KY742343 | KY742174 | |||
| LC 4859 | Camellia sinensis | China | KY742256 | KY742102 | KY742344 | KY742175 | ||||
| E.layuense | CGMCC 3.18362 | T | Perilla sp. | China | KY742261 | KY742107 | KY742349 | – | ||
| LC 8156 | Perilla sp. | China | KY742262 | KY742108 | KY742350 | – | ||||
| E.nigrum | CBS 125.82 | Human toe nail | The Netherlands | GU237974 | FJ426995 | FJ427106 | KT389631 | |||
| CBS 173.73 | T | Dactylis glomerata | USA | GU237975 | FJ426996 | FJ427107 | KT389632 | |||
| E.ovisporum | Epicoccum sorghinum | CBS 180.80 | FMR 14910 | T | Zea mays | South Africa | LT623212 | FJ427068 | FJ427174 | LT623252 |
| E.pimprinum | PD 77/1028 | Soil | India | GU237977 | FJ427050 | FJ427160 | KT389633 | |||
| E.plurivorum | CBS 558.81 | FMR 14909 | T | Setaria sp. | New Zealand | GU238132 | GU237888 | GU237647 | KT389634 | |
| E.pneumoniae | Epicoccum sorghinum | UTHSC:DI16-257; FMR 13747 | T | Human respiratory tract | USA | LN907400 | LT592927 | LT592996 | LT593065 | |
| E.poae | LC 8161 | Poa annua | USA | KY742268 | KY742114 | KY742356 | KY742183 | |||
| CGMCC 3.18363 | T | Poa annua | USA | KY742267 | KY742113 | KY742355 | KY742182 | |||
| LC 8162 | Poa annua | USA | KY742269 | KY742115 | KY742357 | KY742184 | ||||
| E.proteae | Phoma proteae | CBS 114179 | CPC 1854; FMR 15332 | T | Protea cv. carnival | South Africa | JQ044452 | JQ044433 | LT623230 | LT623251 |
| E.sorghinum | CBS 179.80 | Sorghum vulgare | Puerto Rico | GU237978 | FJ427067 | FJ427173 | KT389635 | |||
| CBS 627.68 | Citrus sp. | France | GU237979 | FJ427072 | FJ427178 | KT389636 | ||||
| UTHSC:DI16-288; FMR 13780 | Human superficial tissue | USA | LN907431 | LT592940 | LT593009 | LT593079 | ||||
| UTHSC:DI16-301; FMR 13794 | Human respiratory tract | USA | LN907444 | LT592948 | LT593017 | LT593087 | ||||
| E.viticis | BRIP 29294; LC 5257 | Andropogon gayanus | Australia | KY742271 | KY742117 | KY742359 | – | |||
| CGMCC 3.18344 | T | Vitex negundo | China | KY742272 | KY742118 | KY742360 | KY742186 | |||
| Foliophoma fallens | CBS 161.78 | Olea europaea | New Zealand | GU238074 | KY929147 | – | KC584502 | |||
| CBS 284.70 | Nerium oleander | Italy | GU238078 | KY929148 | – | – | ||||
| Halojulella avicenniae | BCC 18422 | Mangrove wood | Thailand | GU371823 | – | – | GU371787 | |||
| BCC 20173 | Mangrove wood | Thailand | GU371822 | – | – | GU371786 | ||||
| Heterophoma adonidis | CBS 114309 | Adonis vernalis | Sweden | KT389724 | KT389506 | KT389803 | KT389637 | |||
| H.nobilis | CBS 507.91 | Dictamnus albus | The Netherlands | GU238065 | GU237877 | GU237603 | KT389638 | |||
| H.verbascicola | CGMCC 3.18364 | T | Verbascum thapsus | China | KY742273 | KY742119 | KY742361 | KY742187 | ||
| LC 8164 | Verbascum thapsus | China | KY742274 | KY742120 | KY742362 | KY742188 | ||||
| Juxtiphoma eupyrena | Phoma eupyrena | CBS 374.91 | FMR 15329 | Solanum tuberosum | The Netherlands | GU238072 | FJ426999 | FJ427110 | LT623268 | |
| Phoma eupyrena | CBS 527.66 | FMR 15337 | Wheat field soil | Germany | GU238073 | FJ427000 | FJ427111 | LT623269 | ||
| Leptosphaeria conoidea | CBS 616.75 | Lunaria annua | The Netherlands | JF740279 | JF740201 | KT389804 | KT389639 | |||
| L.doliolum | CBS 505.75 | T | Urtica dioica | The Netherlands | GQ387576 | JF740205 | JF740144 | KT389640 | ||
| Leptosphaerulina americana | CBS 213.55 | Trifolium pratense | USA | GU237981 | GU237799 | GU237539 | KT389641 | |||
| L.australis | CBS 317.83 | Eugenia aromatica | Indonesia | EU754166 | GU237829 | GU237540 | GU371790 | |||
| Libertasomyces myopori | CBS 141302 | CPC 27354 | T | Myoporum serratum | South Africa | KX228332 | NR_145200 | – | – | |
| L.platani | CBS 142112 | CPC 29609 | T | Platanus sp. | New Zealand | KY173507 | KY173416 | KY173604 | KY173585 | |
| L.quercus | CBS 134.97 | INIFAT C96/108 | T | Quercus ilex | Spain | DQ377883 | – | – | – | |
| Macroventuria anomochaeta | CBS 525.71 | T | Decayed canvas | South Africa | GU237984 | GU237881 | GU237544 | GU456346 | ||
| M.wentii | CBS 526.71 | T | Plant litter | USA | GU237986 | GU237884 | GU237546 | KT389642 | ||
| Microsphaeriopsis olivacea | CBS 233.77 | Pinus laricio | France | GU237988 | GU237803 | GU237549 | KT389643 | |||
| M.proteae | CBS 111319 | CPC 1425 | Protea nitida | South Africa | JN712563 | JN712497 | – | JN712650 | ||
| Neoascochyta argentina | CBS 112524 | T | Triticum aestivum | Argentina | KT389742 | KT389524 | KT389822 | – | ||
| N.cylindrispora | Ascochyta sp. | UTHSC:DI16-352; FMR 13845 | Human superficial tissue | USA | LN907495 | LT592962 | LT593031 | LT593101 | ||
| Ascochyta sp. | CBS 142456 | UTHSC:DI16-359; FMR 13852 | T | Human superficial tissue | USA | LN907502 | LT592963 | LT593032 | LT593102 | |
| N.desmazieri | CBS 297.69 | T | Lolium perenne | Germany | KT389726 | KT389508 | KT389807 | KT389644 | ||
| Ascochyta sp. | UTHSC:DI16-207; FMR 13697 | Human respiratory tract | USA | LN907350 | LT592907 | LT592976 | LT593045 | |||
| Ascochyta sp. | UTHSC:DI16-320; FMR 13813 | Unknown | USA | LN907463 | LT592956 | LT593025 | LT593095 | |||
| Ascochyta sp. | UTHSC:DI16-332; FMR 13825 | Human superficial tissue | USA | LN907475 | LT592958 | LT593027 | LT593097 | |||
| Ascochyta sp. | UTHSC:DI16-341; FMR 13834 | Human superficial tissue | USA | LN907484 | LT592960 | LT593029 | LT593099 | |||
| N.europaea | CBS 820.84 | T | Hordeum vulgare | Germany | KT389729 | KT389511 | KT389809 | KT389646 | ||
| N.exitialis | CBS 118.40 | Unknown | Unknown | KT389732 | KT389514 | KT389812 | KT389647 | |||
| CBS 389.86 | Triticum aestivum | Switzerland | KT389733 | KT389515 | KT389813 | KT389648 | ||||
| N.graminicola | CBS 301.69 | Lolium multiflorum | Germany | KT389737 | KT389519 | KT389817 | KT389650 | |||
| N.graminicola | CBS 816.84 | Hordeum vulgare | Germany | KT389741 | KT389523 | KT389821 | KT389651 | |||
| N.paspali | CBS 560.81 | T | Paspalum dilatatum | New Zealand | GU238124 | FJ427048 | FJ427158 | KP330426 | ||
| N.soli | LC 8166 | Soil | China | KY742276 | KY742122 | KY742364 | – | |||
| CGMCC 3.18365 | T | Soil | China | KY742275 | KY742121 | KY742363 | – | |||
| N.tardicrescens | Neoascochyta sp. | CBS 689.97 | FMR 15352 | T | Hay | Norway | KT389744 | KT389526 | KT389824 | KT389654 |
| Ascochyta sp. | UTHSC:DI16-291; FMR 13783 | Human superficial tissue | USA | LN907434 | LT592942 | LT593011 | LT593081 | |||
| N.triticicola | CBS 544.74 | T | Triticum aestivum | South Africa | EU754134 | GU237887 | GU237488 | KT389652 | ||
| Neocamarosporium betae | CBS 109410 | PD 77/113 | Beta vulgaris | Unknown | EU754178 | KY940790 | – | GU371774 | ||
| CBS 523.66 | IHEM 3915; PD 66/270 | Beta vulgaris | The Netherlands | EU754179 | FJ426981 | KT389842 | KT389670 | |||
| N.calvescens | CBS 246.79 | PD 77/655 | Atriplex hastata | Germany | EU754131 | KY940774 | – | KC584500 | ||
| N.goegapense | CBS 138008 | CPC 23676 | T | Mesembryanthemum sp. | South Africa | KJ869220 | KJ869163 | – | – | |
| Neocucurbitaria aquatica | Pyrenochaeta quercina | CBS 297.74 | FMR 14867 | T | Sea water | Montenegro | EU754177 | LT623221 | LT623238 | LT623278 |
| N.cava | Pyrenochaeta cava | CBS 115979 | FMR 15333 | Unknown | The Netherlands | EU754198 | AY853248 | LT623234 | LT623273 | |
| Pyrenochaeta cava | CBS 257.68 | FMR 15747; IMI 331911 | T | Wheat-field soil | Germany | EU754199 | JF740260 | KT389844 | LT717681 | |
| N.hakeae | Pyrenochaeta hakeae | CBS 142109 | CPC 28920 | T | Hakea sp. | Australia | KY173526 | KY173436 | KY173613 | KY173593 |
| N.irregularis | Pyrenochaeta unguis-hominis | CBS 142791 | UTHSC:DI16-229; FMR 13719 | T | Human subcutaneous tissue | USA | LN907372 | LT592916 | LT592985 | LT593054 |
| N.keratinophila | Pyrenochaeta keratinophila | CBS 121759 | FMR 9444 | T | Man corneal scrapings | Spain | LT623215 | EU885415 | LT623236 | LT623275 |
| N.quercina | Pyrenochaeta quercina | CBS 115095 | FMR 14868 | T | Quercus robur | Italy | GQ387619 | LT623220 | LT623237 | LT623277 |
| N.unguis-hominis | Pyrenochaeta unguis-hominis | UTHSC:DI16-213; FMR 13703 | Unknown | USA | LN907356 | LT592910 | LT592979 | LT593048 | ||
| Pyrenochaeta unguis-hominis | CBS 111112 | FMR 14866 | Agapornis sp. Lung | The Netherlands | GQ387623 | LT623222 | LT623239 | LT623279 | ||
| Pyrenochaeta unguis-hominis | CBS 112.79 | FMR 15748 | Air sample | Wales | GQ387622 | LT717672 | LT717675 | LT717682 | ||
| Neodidymelliopsis achlydis | CBS 256 77 | T | Achlys triphylla | Canada | KT389749 | KT389531 | KT389829 | – | ||
| N.cannabis | CBS 234.37 | Cannabis sativa | Unknown | GU237961 | GU237804 | GU237523 | KP330403 | |||
| N.longicolla | Phoma sp. | UTHSC:DI16-322; FMR 13815 | Human respiratory tract | USA | LN907465 | LT592957 | LT593026 | LT593096 | ||
| CBS 382 96 | T | Soil in desert | Israel | KT389750 | KT389532 | KT389830 | – | |||
| N.polemonii | CBS 109181 | T | Polemonium caeruleum | The Netherlands | GU238133 | GU237746 | KT389828 | KP330427 | ||
| N.xanthina | CBS 383.68 | T | Delphinium sp. | The Netherlands | GU238157 | GU237855 | KT389831 | KP330431 | ||
| Neomicrosphaeriopsis italica | MFLUCC 15-0485 | T | Tamarix sp. | Italy | KU729854 | KU900318 | – | KU674820 | ||
| MFLUCC 15-0484 | Tamarix sp. | Italy | KU729853 | KU900319 | KX453298 | KU695539 | ||||
| Neophaeosphaeria agaves | CBS 136429 | CPC 21264 | T | Agave tequilana var. azul | Mexico | KF777227 | NR_137833 | – | – | |
| N.filamentosa | CBS 102202 | Yucca rostrata | Mexico | GQ387577 | JF740259 | – | GU371773 | |||
| Neoplatysporoides aloicola | CBS 139901 | CPC 24435 | T | Aloe sp. | Tanzania | KR476754 | KR476719 | – | – | |
| Neopyrenochaeta acicola | Pyrenochaeta acicola | CBS 812.95 | FMR 14872 | T | Waterpipe | The Netherlands | GQ387602 | LT623218 | LT623232 | LT623271 |
| N.fragariae | Pyrenochaeta acicola | CBS 101634 | FMR 14871 | T | Fragaria ananassa | The Netherlands | GQ387603 | LT623217 | LT623231 | LT623270 |
| N.inflorescentiae | Pyrenochaeta inflorescentiae | CBS 119222 | FMR 15334 | T | Protea neriifolia | South Africa | EU552153 | EU552153 | LT623233 | LT623272 |
| N.telephoni | Pyrenochaeta telephoni | CBS 139022 | FMR 15754 | T | Screen of a mobile phone | India | KM516290 | KM516291 | LT717678 | LT717685 |
| Neopyrenochaetopsis hominis | Pyrenochaeta sp. | CBS 143033 | UTHSC:DI16-238; FMR 13728 | T | Human superficial tissue | USA | LN907381 | LT592923 | LT592992 | LT593061 |
| Nothophoma anigozanthi | CBS 381.91 | FMR 14914 | T | Anigozanthus maugleisii | The Netherlands | GU238039 | GU237852 | GU237580 | KT389655 | |
| N.arachidis-hypogaeae | CBS 125.93 | Arachis hypogaea | India | GU238043 | GU237771 | GU237583 | KT389656 | |||
| N.gossypiicola | CBS 377.67 | FMR 14912 | Gossypium sp. | USA | GU238079 | GU237845 | GU237611 | KT389658 | ||
| Phoma sp. | UTHSC:DI16-294; FMR 13787 | Human deep tissue/fluids | USA | LN907437 | LT592943 | LT593012 | LT593082 | |||
| N.infossa | CBS 123395 | T | Fraxinus pennsylvanica | Argentina | GU238089 | FJ427025 | FJ427135 | KT389659 | ||
| N.macrospora | CBS 140674 | UTHSC:DI16-276; FMR 13767 | T | Human respiratory tract | USA | LN880537 | LN880536 | LN880539 | LT593073 | |
| N.quercina | CBS 633.92 | FMR 14913 | Quercus sp. | Ukraine | EU754127 | GU237900 | GU237609 | KT389657 | ||
| Leptosphaerulina sp. | UTHSC:DI16-270; FMR 13761 | Human superficial tissue | USA | LN907413 | LT592929 | LT592998 | LT593067 | |||
| N.variabilis | Phoma sp. | CBS 142457 | UTHSC:DI16-285; FMR 13777 | T | Human respiratory tract | USA | LN907428 | LT592939 | LT593008 | LT593078 |
| Ochrocladosporium elatum | CBS 146.33 | IMI 049629; ATCC 11280 | Wood pulp | Sweden | EU040233 | EU040233 | – | – | ||
| O.frigidarii | CBS 103.81 | T | Cooled room | Germany | EU040234 | EU040234 | – | – | ||
| Ophiosphaerella herpotricha | CBS 620.86 | AFTOL-ID 1569 | Bromus erectus | Switzerland | DQ678062 | KF498728 | – | DQ677958 | ||
| Paraboeremia adianticola | CBS 187.83 | FMR 15344 | Polystichum adiantiforme | USA | GU238035 | GU237796 | GU237576 | KP330401 | ||
| P.camelliae | CGMCC 3.18106 | T | Camellia sp. | China | KX829042 | KX829034 | KX829058 | KX829050 | ||
| CGMCC 3.18107 | Camellia sp. | China | KX829043 | KX829035 | KX829059 | KX829051 | ||||
| CGMCC 3.18108 | Camellia sp. | China | KX829044 | KX829036 | KX829060 | KX829052 | ||||
| P.litseae | CGMCC 3.18109 | T | Litsea sp. | China | KX829037 | KX829029 | KX829053 | KX829045 | ||
| CGMCC 3.18110 | Litsea sp. | China | KX829038 | KX829030 | KX829054 | KX829046 | ||||
| P.oligotrophica | CGMCC 3.18111 | T | Limestone | China | KX829039 | KX829031 | KX829055 | KX829047 | ||
| CGMCC 3.18112 | Limestone | China | KX829040 | KX829032 | KX829056 | KX829048 | ||||
| P.putaminum | CBS 130.69 | FMR 15338 | Malus sylvestris | Denmark | GU238138 | GU237777 | GU237652 | LT623254 | ||
| P.selaginellae | CBS 122.93 | FMR 15348 | T | Selaginella sp. | The Netherlands | GU238142 | GU237762 | GU237656 | LT623255 | |
| Paraconiothyrium estuarinum | CBS 109850 | FMR 14887 | T | Sediment from estuarine | Brazil | JX496129 | JX496016 | JX496355 | LT854937 | |
| Paracucurbitaria italica | Pyrenochaeta corni | CBS 234.92 | FMR 14869 | T | Olea europaea | Italy | EU754176 | LT623219 | LT623235 | LT623274 |
| P.corni | Pyrenochaeta corni | CBS 248.79 | FMR 16593 | Fraxinus excelsior | The Netherlands | GQ387608 | LT903672 | LT900365 | LT903673 | |
| Paraepicoccum amazonense | MFLUCC 15-0493 | Tamarix sp. | Italy | KU900294 | KU752190 | – | KU820871 | |||
| MFLUCC 15-0491 | Tamarix sp. | Italy | KU900295 | KU752191 | – | KU820872 | ||||
| Paraleptosphaeria dryadis | CBS 643.86 | ETH 9446 | Dryas octopetala | Switzerland | GU301828 | JF740213 | – | GU371733 | ||
| Parapyrenochaeta acaciae | Pyrenochaeta acaciae | CBS 141291 | FMR 15755; CPC 25527 | T | Acacia sp. | Australia | KX228316 | KX228265 | LT717679 | LT717686 |
| P.protearum | Pyrenochaeta protearum | CBS 131315 | FMR 15752; CPC 18322 | T | Protea mundii | South Africa | JQ044453 | JQ044434 | LT717677 | LT717683 |
| Pyrenochaeta pinicola | CBS 137997 | FMR 15753; CPC 23455 | Pinus sp. | France | KJ869209 | KJ869152 | KJ869249 | LT717684 | ||
| Phaeomycocentrospora cantuariensis | CBS 132014 | CPC 11694 | Humulus scandens | South Korea | GU253716 | GU269668 | – | – | ||
| CPC 10157 | Humulus scandens | South Korea | GU253712 | GU269664 | – | – | ||||
| Phaeosphaeria oryzae | CBS 110110 | T | Oryza sativa | Korea | KF251689 | KF251186 | KF252680 | – | ||
| Phoma herbarum | CBS 502.91 | Nerium sp. | The Netherlands | GU238082 | GU237874 | GU237613 | KP330419 | |||
| CBS 615.75 | FMR 15340 | Rosa multiflora cv. Cathayensis | The Netherlands | KF251715 | FJ427022 | KF252703 | KP330420 | |||
| CBS 377.92 | Human leg | UK | KT389756 | KT389536 | KT389837 | KT389663 | ||||
| UTHSC:DI16-319; FMR 13812 | Human superficial tissue | USA | LN907462 | LT592955 | LT593024 | LT593024 | ||||
| UTHSC:DI16-204; FMR 13694 | Human deep tissue/fluids | USA | LN907347 | LT592904 | LT592973 | LT593042 | ||||
| UTHSC:DI16-212; FMR 13702 | Human respiratory tract | USA | LN907355 | LT592909 | LT592978 | LT593047 | ||||
| UTHSC:DI16-306; FMR 13799 | Human respiratory tract | USA | LN907449 | LT592952 | LT593021 | LT593091 | ||||
| UTHSC:DI16-307; FMR 13800 | Human respiratory tract | USA | LN907450 | LT592953 | LT593022 | LT593092 | ||||
| Phomatodes aubrietiae | CBS 627.97 | T | Aubrietia sp. | The Netherlands | GU238045 | GU237895 | GU237585 | KT389665 | ||
| P.nebulosa | CBS 100191 | Thlaspi arvense | Poland | KP330446 | KP330434 | KP330390 | KT389666 | |||
| CBS 740.96 | Armoracia rusticana | The Netherlands | KT389758 | KT389540 | KT389839 | KT389667 | ||||
| Pleiochaeta ghindensis | CBS 552.92 | Acacia mellifera | Namibia | EU167561 | EU167561 | – | – | |||
| P.setosa | CBS 496.63 | MUCL 8091 | Cytisus racemosus | Germany | EU167563 | EU167563 | – | – | ||
| Pleospora herbarum | CBS 191.86 | T | Medicago sativa | Uttar Pradesh | JX681120 | NR_111243 | – | KC584471 | ||
| P.typhicola | CBS 132.69 | Typha angustifolia | The Netherlands | JF740325 | – | KT389843 | KC584505 | |||
| Preussia terricola | AFTOL-ID 282; DAOM 230091 | Unknown | Unknown | AY544686 | KT225529 | – | DQ470895 | |||
| Pseudoascochyta novae-zelandiea | CBS 141689 | FMR 15110; ICMP 10493 | T | Cordyline australis | New Zealand | LT592893 | LT592892 | LT592894 | LT592895 | |
| P.pratensis | CBS 141688 | FMR 14524 | T | Soil | Spain | LT223131 | LT223130 | LT223132 | LT223133 | |
| Pseudopyrenochaeta lycopersici | Pyrenochaeta lycopersici | CBS 306.65 | FMR 15746 | T | Lycopersicon esculentum | Germany | EU754205 | NR_103581 | LT717674 | LT717680 |
| P.terretris | Pyrenochaeta lycopersici | CBS 282.72 | FMR 15327 | T | Soil | The Netherlands | LT623216 | LT623228 | LT623246 | LT623287 |
| Pyrenochaeta nobilis | CBS 407.76 | FMR 14870 | T | Laurus nobilis | Italy | EU754206 | EU930011 | KT389845 | LT623276 | |
| Pyrenochaetopsis americana | Pyrenochaetopsis sp. | UTHSC:DI16-225; FMR 13715 | Unknown | USA | LN907368 | LT592912 | LT592981 | LT593050 | ||
| P.botulispora | Pyrenochaetopsis sp. | UTHSC:DI16-289; FMR 13781 | Human respiratory tract | USA | LN907432 | LT592941 | LT593010 | LT593080 | ||
| Pyrenochaetopsis sp. | UTHSC:DI16-297; FMR 13790 | Human superficial tissue | USA | LN907440 | LT592945 | LT593014 | LT593084 | |||
| Pyrenochaetopsis sp. | CBS 142458 | UTHSC:DI16-298; FMR 13791 | T | Human respiratory tract | USA | LN907441 | LT592946 | LT593015 | LT593085 | |
| P.confluens | Pyrenochaetopsis sp. | CBS 142459 | UTHSC:DI16-303; FMR 13796 | T | Human deep tissue/fluids | USA | LN907446 | LT592950 | LT593019 | LT593089 |
| P.decipiens | CBS 343.85 | FMR 14880 | T | Globodera pallida | The Netherlands | GQ387624 | LT623223 | LT623240 | LT623280 | |
| P.globosa | Pyrenochaetopsis sp. | CBS 143034 | UTHSC:DI16-275; FMR 13766 | T | Human superficial tissue | USA | LN907418 | LT592934 | LT593003 | LT593072 |
| P.indica | CBS 124454 | FMR 14879 | T | Saccharum officinarum | India | GQ387626 | LT623224 | LT623241 | LT623281 | |
| P.leptospora | CBS 101635 | FMR 14877 | T | Secale cereale | Unknow | GQ387627 | JF740262 | LT623242 | LT623282 | |
| Coniothyrium cereale | CBS 122787 | FMR 14873 | Unknown | Germany | EU754151 | LT623225 | LT623243 | LT623283 | ||
| P.microspora | Pyrenochaetopsis sp. | UTHSC:DI16-198; FMR 13688 | Human superficial tissue | USA | LN907341 | LT592899 | LT592968 | LT593037 | ||
| CBS 102876 | FMR 14874 | T | Water | Montenegro | GQ387631 | LT623226 | LT623244 | LT623284 | ||
| P.paucisetosa | Pyrenochaetopsis sp. | CBS 142460 | UTHSC:DI16-193; FMR 13683 | T | Human superficial tissue | USA | LN907336 | LT592897 | LT592966 | LT593035 |
| P.poae | CBS 136769 | FMR 14876 | T | Poa sp. | The Netherlands | KJ869175 | KJ869117 | KJ869243 | LT623286 | |
| P.setosissima | Pyrenochaetopsis microspora | CBS 119739 | FMR 14875 | T | Coffea arabica | Brazil | GQ387632 | LT623227 | LT623245 | LT623285 |
| P.tabarestanensis | CBS 139506 | IBRC-M 30051 | T | Soil | Iran | KF803343 | KF730241 | KX789523 | – | |
| P.uberiformis | Pyrenochaetopsis sp. | CBS 142461 | UTHSC:DI16-277; FMR 13769 | T | Human superficial tissue | USA | LN907420 | LT592935 | LT593004 | LT593074 |
| Querciphoma carteri | CBS 105.91 | Quercus robur | Germany | KF251712 | JF740181 | KF252700 | KT389591 | |||
| Remotididymella anthropophila | Phoma sp. | CBS 142462 | UTHSC:DI16-278; FMR 13770 | T | Human respiratory tract | USA | LN907421 | LT592936 | LT593005 | LT593075 |
| R.destructiva | Phoma destructiva var. destructiva | CBS 133.93 | FMR 15349 | Solanum lycopersicon | Guadeloupe | GU238064 | GU237779 | GU237602 | LT623257 | |
| Phoma destructiva var. destructiva | CBS 378.73 | FMR 15328 | T | Lycopersicon esculentum | Tonga | GU238063 | GU237849 | GU237601 | LT623258 | |
| Phoma destructiva var. diversispora | CBS 162.78 | FMR 14906 | Lycopersicon esculentum | The Netherlands | GU238062 | GU237788 | GU237600 | LT623259 | ||
| Shiraia bambusicola | NBRC 30754 | Phyllostachys sp. | Japan | AB354969 | AB354988 | AB355003 | – | |||
| NBRC 30771 | Phyllostachys sp. | Japan | AB354971 | AB354990 | AB355005 | – | ||||
| NBRC 30753 | Phyllostachys sp. | Japan | AB354968 | AB354987 | AB355002 | – | ||||
| NBRC 30772 | Phyllostachys sp. | Japan | AB354972 | AB354991 | AB355006 | – | ||||
| Similiphoma crystallifera | Phoma crystallifera | CBS 193.82 | FMR 15343 | T | Chamaespartium sagittale | Austria | GU238060 | GU237797 | GU237598 | LT623267 |
| Sporormiella minima | CBS 524.50 | AFTOL-ID 1256 | Dung of goat | Panama | DQ678056 | KT389543 | – | DQ677950 | ||
| Stagonosporopsis dorenboschii | CBS 426.90 | T | Physostegia virginiana | The Netherlands | GU238185 | GU237862 | GU237690 | KT389678 | ||
| S.hortensis | CBS 572.85 | Phaseolus vulgaris | The Netherlands | GU238199 | GU237893 | GU237704 | KT389681 | |||
| Staurosphaeria aloes | Hazslinszkyomyces aloes | CBS 136437 | CPC 21572 | T | Aloe dichotoma | South Africa | KF777198 | NR_137821 | – | – |
| S.aptrootii | Hazslinszkyomyces aptrootii | CBS 483.95 | T | Lycium sp. | The Netherlands | GU301806 | KY929149 | – | – | |
| S.lyciicola | Hazslinszkyomyces lycii | CBS 142619 | CPC 30998 | T | Lycium barbarum | Hungary | KY929180 | KY929150 | – | – |
| Hazslinszkyomyces lycii | CPC 31014 | Lycium barbarum | Hungary | KY929181 | KY929151 | – | – | |||
| Vacuiphoma bulgarica | Phoma bulgarica | CBS 357.84 | FMR 14917 | T | Trachystemon orientale | Bulgaria | GU238050 | GU237837 | GU237589 | LT623256 |
| V.oculihominis | Phoma sp. | UTHSC:DI16-308; FMR 13801 | T | Human superficial tissue | USA | LN907451 | LT592954 | LT593023 | LT593093 | |
| Xenodidymella applanata | CBS 205.63 | Rubus idaeus | The Netherlands | GU237998 | GU237798 | GU237556 | KP330402 | |||
| CBS 115577 | Rubus idaeus | Sweden | KT389762 | KT389546 | KT389850 | KT389688 | ||||
| X.asphodeli | CBS 375.62 | T | Asphodelus albus | France | KT389765 | KT389549 | KT389853 | KT389689 | ||
| X.catariae | CBS 102635 | Nepeta catenaria | The Netherlands | GU237962 | GU237727 | GU237524 | KP330404 | |||
| X.humicola | CBS 220.85 | Franseria sp. | USA | GU238086 | GU237800 | GU237617 | KP330422 | |||
| X.saxea | Phoma saxea | CBS 419.92 | FMR 15347 | T | Corroded mediterranean marble | Unknown | GU238141 | GU237860 | GU237655 | KP330429 |
| Xenopyrenochaetopsis pratorum | Pyrenochaetopsis pratorum | CBS 445.81 | FMR 14878 | T | Lolium perenne | New Zealand | GU238136 | JF740263 | KT389846 | KT389671 |
AFTOL: Assembling the Fungal Tree of Life; ATCC: American Type Culture Collection, Virginia, USA; BCC: Biotec Culture Collection, Pathum Thani, Thailand; BCCM/IHEM: Biomedical Fungi and Yeasts Collection, Louvain-la-Neuve, Belgium; BCCM/MUCL: Mycothèque de l'Université catholique de Louvain, Louvain-la-Neuve, Belgium; BRIP: Plant Pathology Herbarium, Department of Employment, Economic, Development and Innovation, Queensland, Australia; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection, Beijing, China; CMW: Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; CPC: Culture collection of Pedro Crous, housed at CBS; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; ETH: Herbaria of the department of Environmental Systems Science. Institute of Integrative Biology, Zürich, Switzerland; FMR, Facultat de Medicina, Universitat Rovira i Virgili, Reus, Spain; IBRC: Iranian Biological Resources Center, Tehran, Iran; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IFO: Institute for Fermentation, Osaka, Japan, now NBRC; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, U.K.; INIFAT: Instituto de Investigaciones Fundamentales en Agricultura Tropical “Alejandro de Humboldt”, Santiago de las Vegas, Cuba; LC: Corresponding author's personal collection deposited in laboratory, housed at CAS, China; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NBRC: Biological Resource Center, National Institute of Technology and Evaluation, Chiba, Japan; PD: Plant Protection Service, Wageningen, the Netherlands; UTHSC, Fungus Testing Laboratory at the University of Texas Health Science Center, San Antonio, Texas, USA.
T: ex-type strain
ITS: internal transcibed spacer regions 1 & 2 including 5.8S nrDNA gene; LSU: 28S large subunit of the nrRNA gene; RPB2: RNA polymerase II second subunit; TUB: ß-tubulin.
Phenotypic study
For cultural characterisation, isolates were grown on oatmeal agar (OA; 30 g of filtered oat flakes, 15 g of agar-agar, 1 L tap water) and malt extract agar (MEA; 40 g of malt extract, 15 g of agar-agar, 1 L distilled water), at 25 ± 1 °C for 14 d in darkness (recipes according to Boerema et al. 2004 and Crous et al. 2009). Some of the cultures were incubated under near-ultraviolet (UV) light (12 h light, 12 h dark) or on carnation leaf agar (CLA) to induce sporulation if necessary (Fisher et al., 1982, Su et al., 2012). Colony diameters were measured after 7 d at 25 ± 1 °C, and colony characterisation was performed 14 d after inoculation on the culture media. Colours were according to Kornerup & Wanscher (1978). The ability of the isolates to grow at cardinal temperatures were determined on potato dextrose agar (PDA; Pronadisa, Madrid, Spain) after 7 d in darkness, ranging from 5 to 35 °C at 5 °C intervals, and including 37 °C.
Micromorphological characterisation was performed by examining at least 30 individuals of each structure (Aveskamp et al., 2010, Chen et al., 2015). Wet mounts (in Shear's mounting medium and in water) of structures were examined by using an Olympus CH2 compound microscope (Olympus Corporation, Tokyo, Japan). Photo micrographs were captured using a Zeiss Axio-Imager M1 microscope (Oberkochen, Germany) with a DeltaPix Infinity X digital camera using Nomarski differential interference contrast. The production of metabolite E+ (NaOH spot test) was carried out by the application of a droplet of 1N NaOH on a colony grown on MEA (Dorenbosch, 1970, Noordeloos et al., 1993).
DNA isolation, PCR amplification and sequencing
The total genomic DNA was extracted from colonies grown on PDA after 7 d incubation at 20 ± 1 °C, using the FastDNA kit protocol (Bio101, Vista, CA), with a FastPrep FP120 instrument (Thermo Savant, Holbrook, NY) according to the manufacturer's protocol. DNA was quantified by using Nanodrop 2000 (Thermo Scientific, Madrid, Spain). The following loci were amplified and sequenced: a fragment of the 28S nrRNA gene (LSU) with the primer pair LR0R (Rehner & Samuels 1994) and LR5 (Vilgalys & Hester 1990), internal transcribed spacer region (ITS1-5.8S-ITS2) with the primer pair ITS5 and ITS4 (White et al. 1990), a fragment of the beta-tubulin gene (tub2) with the primers TUB2Fw and TUB4Rd (Woudenberg et al. 2009) and a fragment of the RNA polymerase II subunit 2 gene (rpb2) with RPB2-5F2 (Sung et al. 2007) and fRPB2-7cR primers (Liu et al. 1999). The PCR amplifications were performed in a total volume of 25 μL containing 5 μL 10× PCR Buffer (Invitrogen, California, USA), 0.2 mM dNTPs, 0.5 μM of each primer, 1 U Taq DNA polymerase and 1–10 ng genomic DNA. PCR conditions for LSU, ITS and tub2 were set as follows: an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation, annealing and extension, and a final extension step at 72 °C for 10 min. For the LSU and ITS amplification, the 35 cycles consisted of 45 s at 95 °C, 45 s at 53 °C and 2 min at 72 °C; and for the tub2 region 30 s at 94 °C, 45 s at 56 °C and 1 min at 72 °C. The PCR program for rpb2 amplification consisted of 5 cycles of 45 s at 94 °C, 45 s at 60 °C and 2 min at 72 °C, then 5 cycles with a 58 °C annealing temperature and 30 cycles with a 54 °C annealing temperature (Woudenberg et al. 2013). Sequencing of the amplicons was made in both directions with the same primer pair used for amplification at Macrogen Europe (Macrogen Inc., Amsterdam, The Netherlands). The consensus sequences were obtained using the SeqMan software v. 7 (DNAStar Lasergene, Madison, WI, USA).
Phylogenetic analyses
Sequences of related species described in previous studies were obtained from GenBank (Aveskamp et al., 2009, Aveskamp et al., 2010, De Gruyter et al., 2010, De Gruyter et al., 2013, Wijayawardene et al., 2014, Chen et al., 2015, Chen et al., 2017, Thambugala et al., 2016), and listed in Table 1. For the phylogenetic study, the alignments of the sequences were performed using MEGA v. 6.06 (Tamura et al. 2013), using the ClustalW application (Thompson et al. 1994), refined with MUSCLE (Edgar 2004) and manually adjusted using the same software platform. The ambiguous regions were excluded from the analyses. Phylogenetic reconstructions were made by maximum-likelihood (ML) and Bayesian inference (BI) with RAxML v. 8.2.10 (Stamatakis 2014) and MrBayes v. 3.2.6 (Ronquist et al. 2012), respectively. The best substitution model for each gene matrix correspond to GTR+I+G, and was estimated using MrModelTest v. 2.3 (Nylander 2004). For ML analyses, nearest-neighbour interchange was used as the heuristic method for tree inference. Support for internal branches was assessed by 1 000 ML bootstrapped pseudoreplicates. Bootstrap support (BS) ≥70 was considered significant. For BI analyses, Markov chain Monte Carlo (MCMC) sampling was performed with 46 M generations, with samples taken every 1 000 generations. The 50 % majority rule consensus trees and posterior probability values (PP) were calculated after removing the first 25 % of the resulting trees for burn-in. A PP value ≥0.95 was considered as significant. Both ML and BS analyses were run in CIPRES (Miller et al. 2012). Preussia terricola (AFTOL-ID 282) and Sporormiella minima (CBS 524.50) served as outgroup taxa. Sequences generated in this study were deposited in GenBank (see Table 1), the final matrices used for phylogenetic analyses in TreeBASE (www.treebase.org; accession number: S21115) and the novel taxonomic descriptions and nomenclature in MycoBank (www.mycobank.org; Crous et al. 2004).
Results
Phylogenetic analyses
The final concatenated dataset obtained with both ML and Bayesian analysis contained 357 ingroup strains with a total of 1 888 characters including gaps (519 for LSU, 336 for ITS, 434 for tub2 and 599 for rpb2), of which 742 are parsimony informative (132 for LSU, 111 for ITS, 149 for tub2 and 350 for rpb2). The sequence datasets did not show conflict in the tree topologies for the 70 % reciprocal bootstrap trees, which allowed to combine the four genes for the multi-locus analysis.
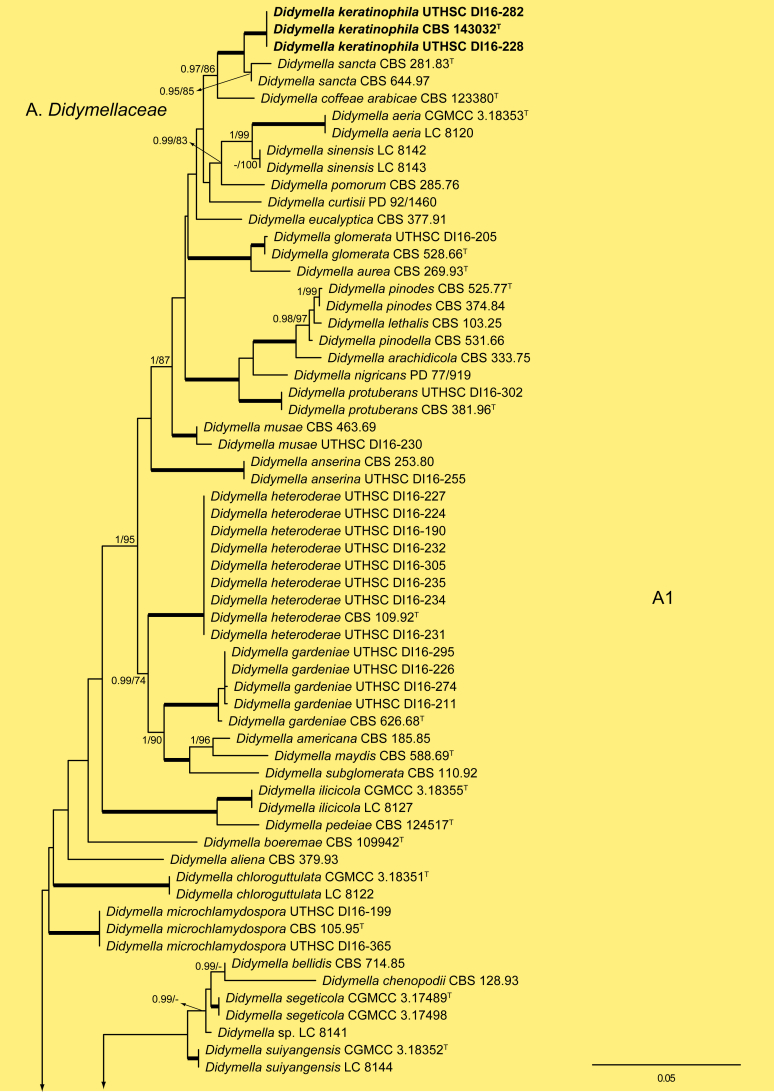
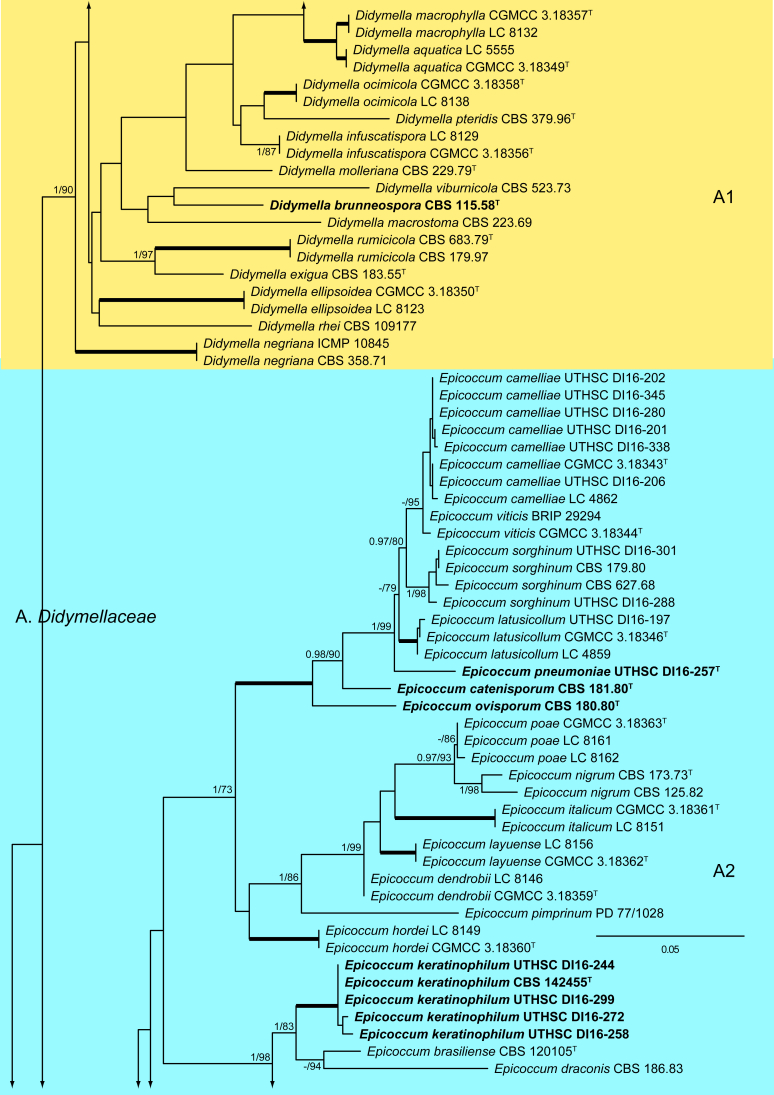
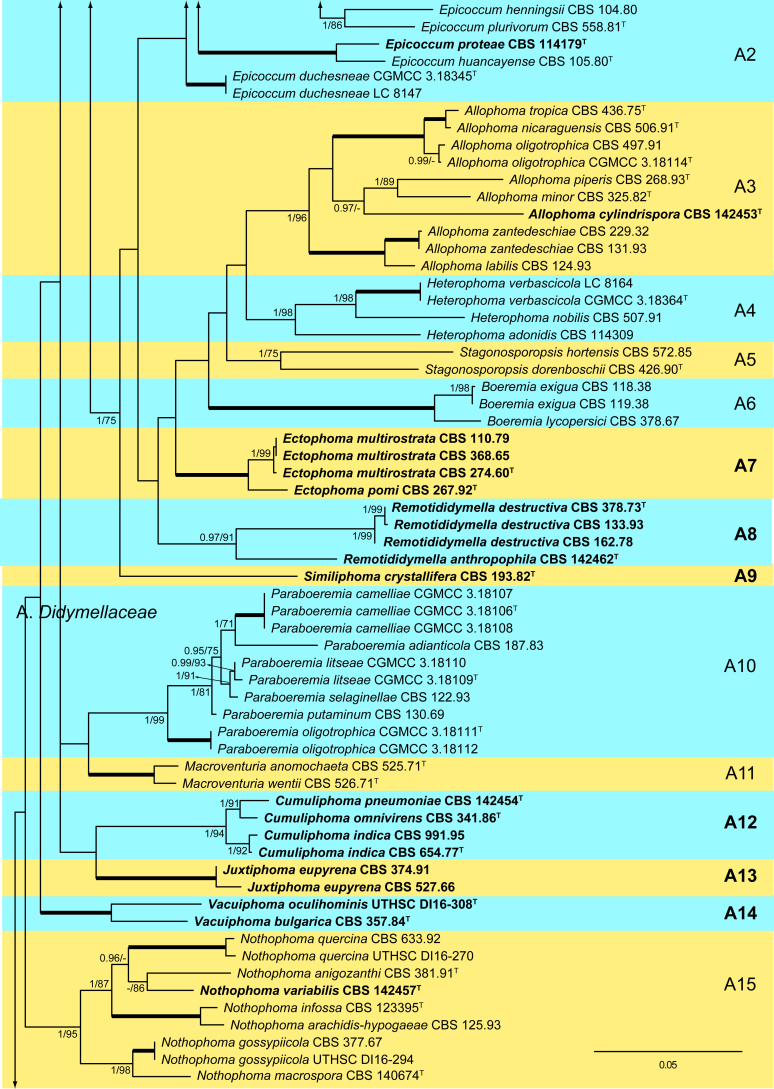
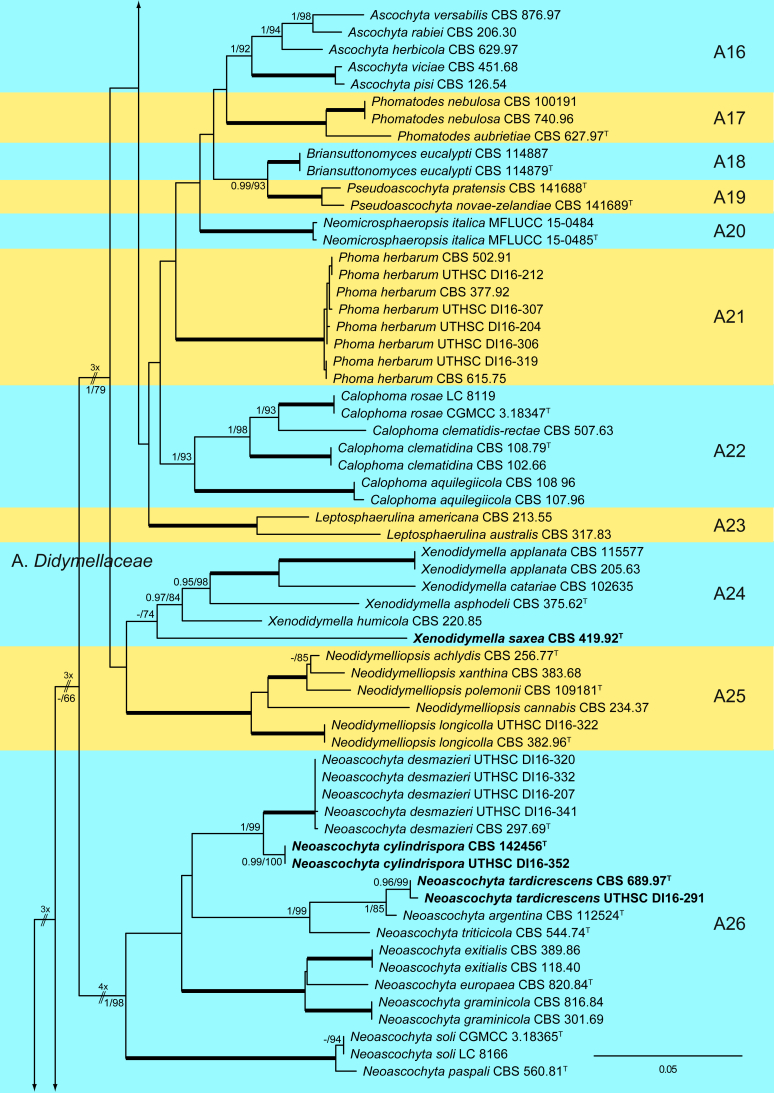
The ML analysis showed similar tree topology and was congruent with that obtained in the Bayesian analysis. For the BI multi-locus analysis, a total of 34 677 trees were sampled after the burn-in with a stop value of 0.01. The support values were slightly different with the two analysis methods; with BI, posterior probabilities being higher than the ML bootstrap support values (Fig. 1).
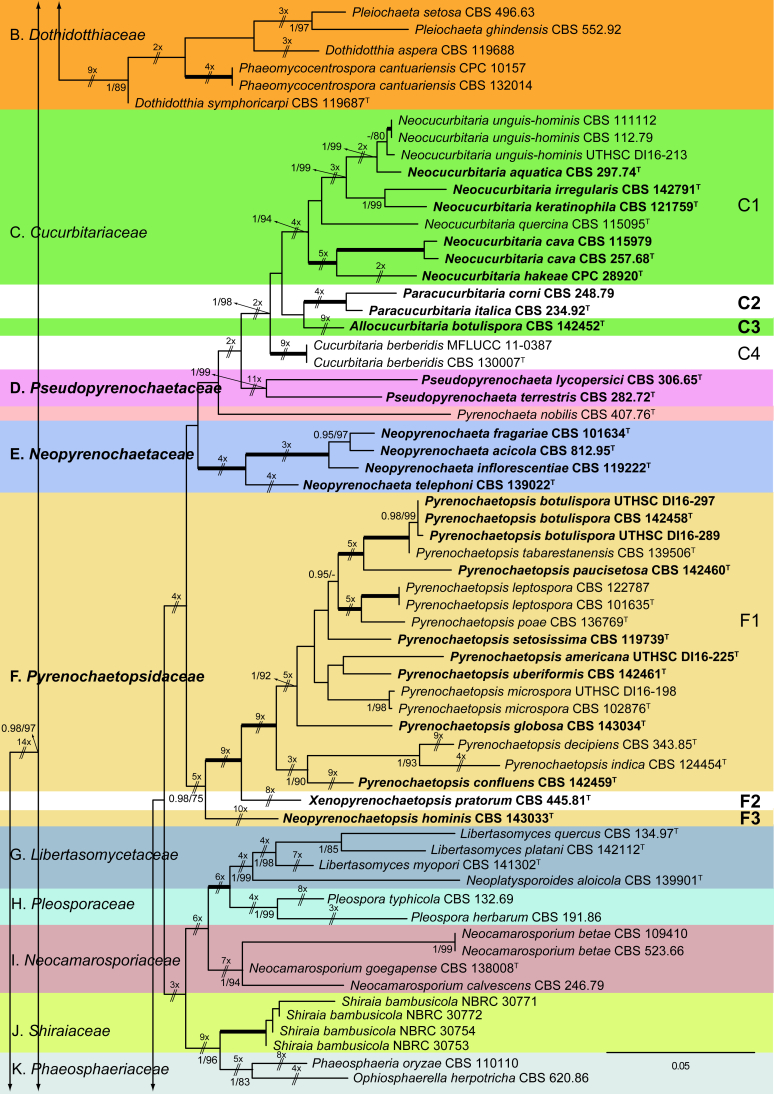
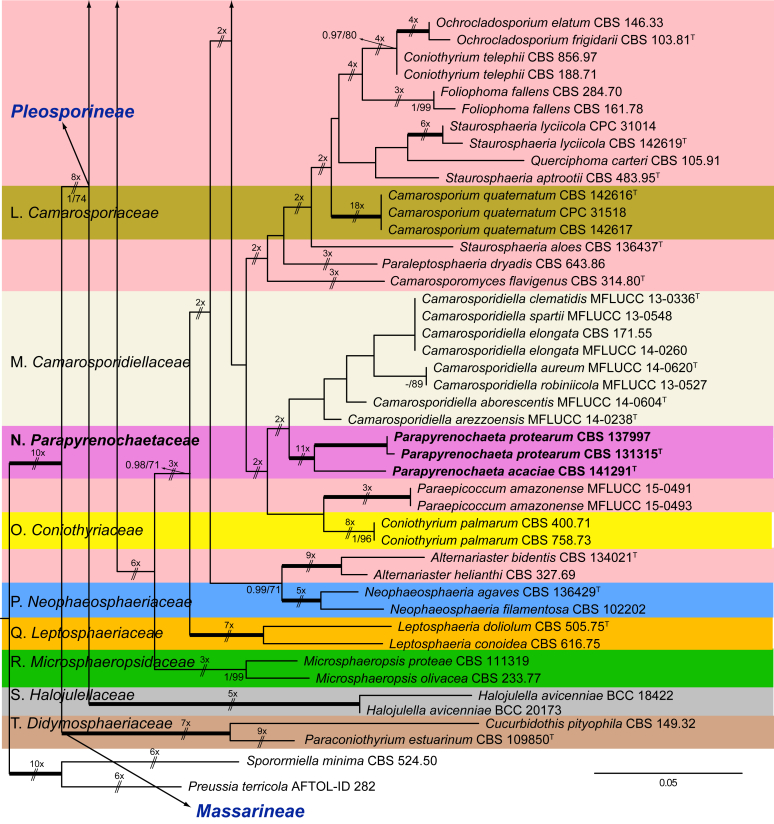
Fig. 1.
Phylogenetic tree inferred from a Maximum likelihood analysis based on a concatenated alignment of LSU, ITS, tub2 and rpb2 sequences of 357 strains representing species in Cucurbitariaceae, Didymellaceae and allied families within Pleosporales. The Bayesian posterior probabilities (PP) above 0.95 and the RAxML bootstrap support values (BS) above 70 % are given at the nodes (PP/BS). Fully supported branched (1 PP/100 BS) are indicated in bold. Some branches were shortened to fit them to the page, these are indicated by two diagonal lines with the number of times a branch was shortened. Newly proposed taxa are given in bold. Type strains are indicated by a superscript T. The tree was rooted with Preussia terricola (AFTOL-ID 828) and Sporormiella minima (CBS 524.50).
The phylogenetic tree distinguished two main supported clades corresponding to the suborders Massarineae (1 PP / 100 % BS), with only the family Didymosphaeriaceae (clade T) here included, and Pleosporineae (1 PP / 74 % BS), emcompassing over 19 families (clades A–S), respectively. Four of the families of the latter suborder are proposed here as new, i.e., Pseudopyrenochaetaceae (clade D), Neopyrenochaetaceae (clade E), Pyrenochaetopsidaceae (clade F) and Parapyrenochaetaceae (clade N). The main clade of the Pleosporineae corresponded to the Didymellaceae (clade A) showing 25 well-supported terminal clades with the only exception being Epicoccum (A2). Twenty terminal clades corresponded to known genera and six are proposed here as new: Ectophoma (A7), Remotididymella (A8), Similiphoma (A9), Cumuliphoma (A12), Juxtiphoma (A13) and Vacuiphoma (A14). The genus Didymella (A1; 1 PP / 90 % BS), comprised 48 species and one undescribed, including two proposed here as new: D. keratinophila sp. nov. (the type strain CBS 143032, UTHSC DI16-228 and UTHSC DI16-282), which is phylogenetically close to D. sancta and D. coffeae-arabicae, and D. brunneospora sp. nov. (CBS 115.58). Several of the clinical strains included in Didymella were distributed among seven known species, i.e., D. heteroderae (nine strains), D. gardeniae (four strains), D. microchlamydospora (two strains), and D. anserina, D. glomerata, D. musae and D. protuberans with only one strain for each. Epicoccum (A2; unsupported) was represented by 17 previously described species (including the type species E. nigrum), the new species E. catenisporum sp. nov., E. ovisporum sp. nov., E. pneumoniae sp. nov. (phylogenetically related with E. camelliae, E. latusicollum, E. sorghinum and E. viticis), and E. keratinophilum sp. nov. (phylogenetically related with E. brasiliense and E. draconis). Finally, E. proteae (basionym Phoma proteae), which clustered with E. huancayense, is here combined in Epicoccum. Allophoma (clade A3; 1 PP / 96 % BS) is enlarged with A. cylindrispora sp. nov., previously identified as Phoma sp. (Valenzuela-Lopez et al. 2016), clustering with A. minor and A. piperis. The clades from A4 to A6 encompassed three genera i.e. Heterophoma (A4; 1 PP / 98 % BS), Stagonosporopsis (A5; 1 PP / 75 % BS) and Boeremia (A6; 1 PP / 100 % BS). The new genus Ectophoma (clade A7; 1 PP / 100 % BS) comprise two new combinations previously included in Phoma, i.e. the generic type E. multirostrata (syn. P. multirostrata) and E. pomi (syn. P. pereupyrena). The new genus Remotididymella (A8; 0.97 PP / 91 % BS) comprised R. destructiva comb. nov. (basionym Phoma destructiva), the type species, and the new species R. anthropophila. For Phoma crystallifera the new monotypic genus Similiphoma (clade A9) and the new combination S. crystallifera are proposed. The clade corresponding to the genus Paraboeremia (clade A10; 1 PP / 99 % BS) included the six accepted species. Macroventuria formed a well-supported clade (A11; 1 PP / 100 % BS) and included the ex-type strains of M. anomochaeta and M. wentii. Cumuliphoma gen. nov. (clade A12; 1 PP / 94 % BS) included C. omnivirens comb. nov. (syn. Phoma omnivirens), C. indica sp. nov. (with two strains previously identified as P. omnivirens) and C. pneumoniae sp. nov., the latter represented by a clinical strain. The proposed new monotypic genus Juxtiphoma (clade A13; 1 PP / 100 % BS), includes two strains of J. eupyrena comb. nov. (basionym Phoma eupryrena). The new genus Vacuiphoma (clade A14; 1 PP / 100 % BS), included the type species V. bulgarica comb. nov. (basionym Phoma bulgarica) and the new species V. oculihominis described from a sterile clinical strain (UTHSC DI16-308). The genus Nothophoma (clade A15; 1 PP / 95 % BS) comprised seven species, including the generic type, N. infossa, and N. variabilis sp. nov., which is based on a clinical strain phylogenetically related with the ex-type strain of N. anigozanthi. The clade corresponding to Ascochyta (clade A16; 1 PP / 92 % BS), grouped five species, including the type species A. pisi. Clade A17 (1 PP / 100 % BS) included the type species of Phomatodes (P. aubrietiae) and two strains of P. nebulosa, the other species of the genus. The Briansuttonomyces clade (A18, 1 PP / 100 % BS), included two strains of the only species of the genus, B. eucalypti. The clade A19 (1 PP / 100 % BS) encompassed the ex-type strains of the two species of Pseudoascochyta, P. novae-zelandiae and P. pratensis. The Neomicrosphaeropsis clade (A20; 1 PP / 100 % BS), contained the type species of the genus, N. italica. In Phoma (A21; 1 PP / 100 % BS) eight strains were grouped, all of them identified as P. herbarum (five from clinical origin and three reference strains). The genus Calophoma (A22; 1 PP / 93 % BS) comprised four species: C. aquilegiicola, C. clematidis-rectae, C. clematidina (type species of the genus) and C. rosae. The clade corresponding to Leptosphaerulina (A23; 1 PP / 100 % BS) contained the two known species, L. americana and L. australis. Xenodidymella (A24; - PP / 74 % BS), grouped the four species of this genus and the new combination Xenodidymella saxea (basionym Phoma saxea), which forms a basal clade with a strain of X. humicola. The clade A25 (1 PP / 100 % BS) included five species of Neodidymelliopsis. Neoascochyta (A26; 1 PP / 98 % BS) represented a basal clade of the Didymellaceae, very distant from the other genera of that family, and grouped 10 species, two of which are here proposed as new: Neoascochyta cylindrispora sp. nov. and Neoascochyta tardicrescens sp. nov.
In the family Cucurbitariaceae (clade C; 1 PP / 98 % BS) analyses resulted in four clades, which we recognise as genera. Neocucurbitaria (C1; 1 PP / 94 % BS), which included two new species, N. irregularis (CBS 142791) and N. aquatica (CBS 297.74), and the three new combinations, N. cava (syn. Pyrenochaeta cava), N. hakeae (basionym Pyrenochaeta hakeae) and N. keratinophila (basionym Pyrenochaeta keratinophila); the new genus Paracucurbitaria (C2; 1 PP / 100 % BS), with two species P. corni comb. nov. (syn. Pyrenochaeta corni) and P. italica sp. nov.; the new genus Allocucurbitaria (clade C3), with the type species A. botulispora sp. nov. Finally, the genus Cucurbitaria (clade C4; 1 PP / 100 % BS) including only the type species, C. berberidis.
Pseudopyrenochaetaceae fam. nov. (clade D; 1 PP / 99 % BS) is introduced to accommodate Pyrenochaeta lycopersici and P. terrestris in the new genus Pseudopyrenochaeta.
The generic type of Pyrenochaeta, Pyrenochaeta nobilis, was phylogenetically distant from the Cucurbitariaceae in our phylogeny, and therefore we consider this species as incertae sedis.
The proposed new family Neopyrenochaetaceae (clade E; 1 PP / 100 % BS) encompassed several taxa previously included in Pyrenochaeta. However, since they were located outside from Cucurbitariaceae s. str. we propose the new genus Neopyrenochaeta, with the new combinations: N. acicola (syn. Pyrenochaeta acicola), N. inflorescentiae (basionym. Pyrenochaeta inflorescentiae) and N. telephoni (basionym Pyrenochaeta telephoni), and the new species N. fragariae.
The new family Pyrenochaetopsidaceae (clade F; 0.98 PP / 75 % BS) grouped three clades, which correspond to the genera Pyrenochaetopsis, the type genus (type species, P. leptospora) (F1; 1 PP / 100 % BS), Xenopyrenochaetopsis (type species, X. pratorum comb. nov.) (F2) and Neopyrenochaetopsis (type species, N. hominis sp. nov.) (F3). Pyrenochaetopsis encompassed seven new species: P. americana, P. botulispora, P. confluens, P. globosa, P. pauciseptata, P. setosissima and P. uberiformis.
The Clade N (1 PP / 100 % BS), which consists of several isolates previously recognised in Pyrenochaeta, is proposed as the new family Parapyrenochaetaceae. Accordingly, the new genus Parapyrenochaeta is proposed for P. acaciae comb. nov. (basionym Pyrenochaeta acaciae), and the type species Parapyrenochaeta protearum comb. nov. (basionym Pyrenochaeta protearum). The strain CBS 137997, previously identified as Pyrenochaeta pinicola, was re-identified as Parapyrenochaeta protearum.
The monospecific genus Paraepicoccum was introduced by Matsushima (1993), later epitypified as Paraepicoccum amazonense by Thambugala et al. (2016) and considered as incertae sedis in Pleosporineae, which is supported by our phylogenetic results.
Taxonomy
After multi-locus sequence analysis of 357 strains distributed among several families within Pleosporineae and the morphological study of 143 strains, in the present paper we propose: four new families, 13 new genera, 28 new species, 20 new combinations, and four typifications. Novel taxa are described and illustrated. Six species proved to be sterile in culture, and therefore are described based on DNA sequence data, following the approach of Chen et al. (2017). Clades and genera are given as they appear in the phylogenetic tree, and species are listed in alphabetical order.
Clade A: Didymellaceae Gruyter et al., Mycol. Res. 113: 516. 2009.
Type genus: Didymella Sacc.
Clade A1: Didymella
Didymella Sacc. ex Sacc., Syll. Fung. 1: 545. 1882. emend. Chen et al., Stud. Mycol. 82: 173. 2015.
Synonym: Peyronellaea Goid. ex Togliani, Ann. Sperim. Agrar. II 6: 93. 1952.
Type species: Didymella exigua (Niessl) Sacc.
Didymella anserina (Marchal) Q. Chen & L. Cai, Stud. Mycol. 82: 173. 2015.
Basionym: Phoma anserina Marchal, Champignon Copr. 11: 1891.
Synonyms: Peyronellaea anserina (Marchal) Aveskamp et al., Stud. Mycol. 65: 31. 2010.
Phoma radicis-callunae R.W. Rayner, Bot. Gaz. 73: 231. 1922.
Phoma suecica J.F.H. Beyma, Antonie van Leeuwenhoek 8: 110. 1942.
Description: de Gruyter & Noordeloos (1992).
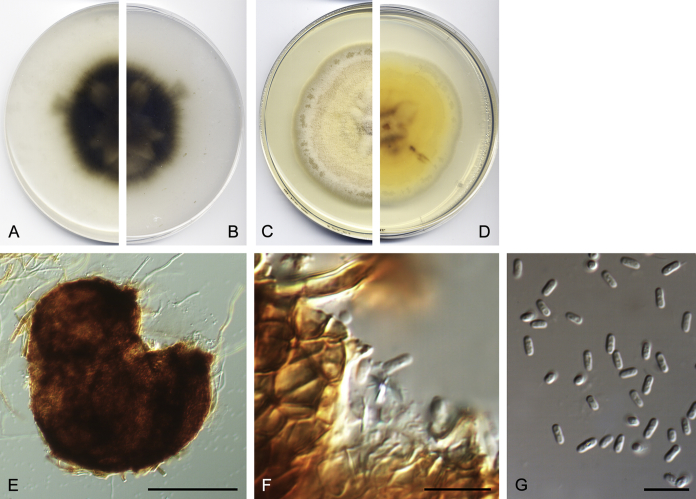
Materials examined: Germany, Giessen, Dec. 1979, R. Hadlok, living culture CBS 253.80. USA, from human sputum sample, 2008, D.A. Sutton, living cultures UTHSC DI16-255 = FMR 13745.
Notes: Didymella anserina is a ubiquitous soil fungus that has been found in Africa, Europe and North America. Although frequently present on herbaceous or woody plants, it has been recorded from many other substrates. Our strain UTHSC DI16-255 is the first report from a human clinical specimen, and it is morphologically similar to the reference strain of D. anserina (CBS 253.80).
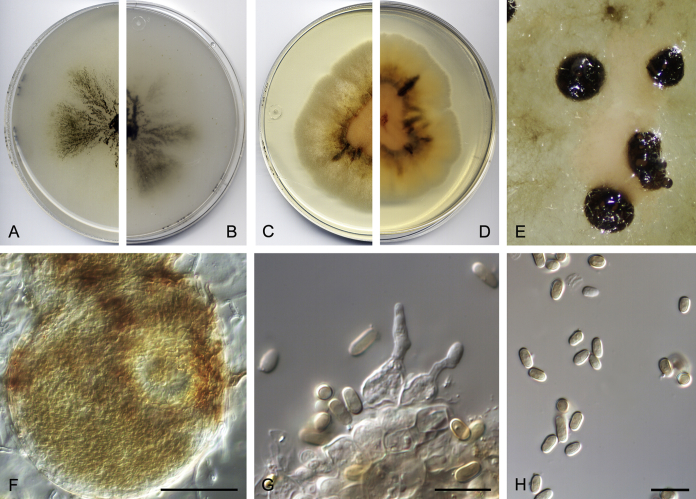
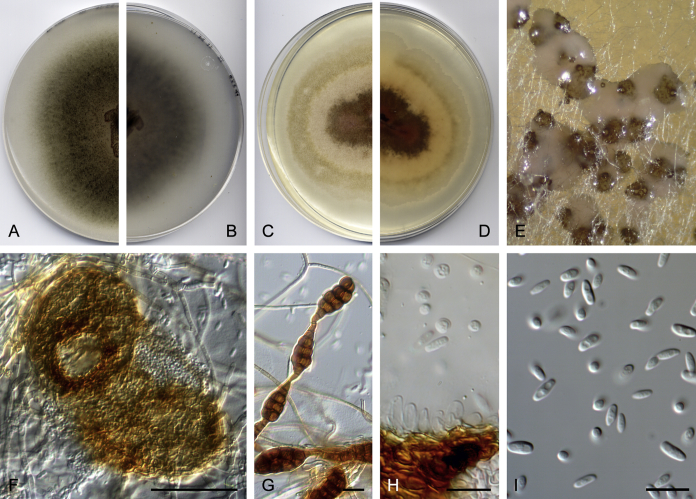
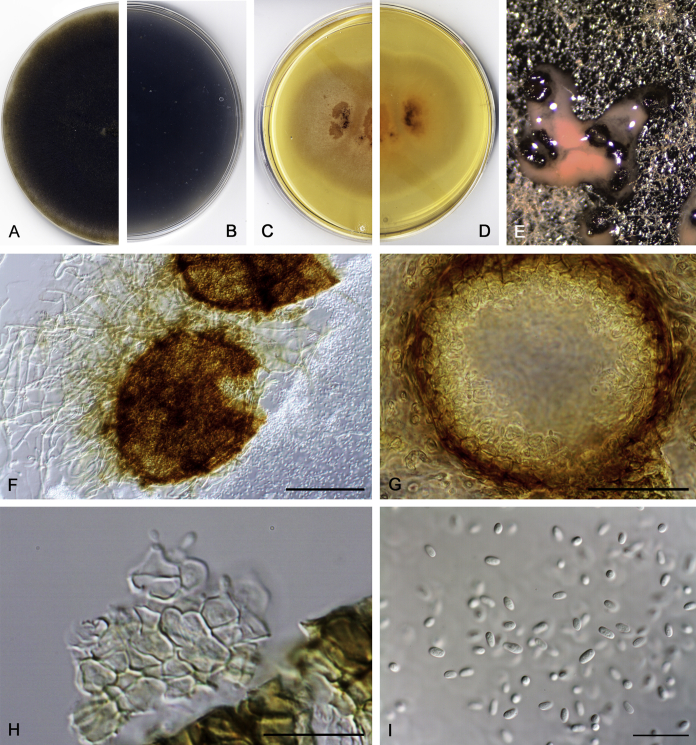
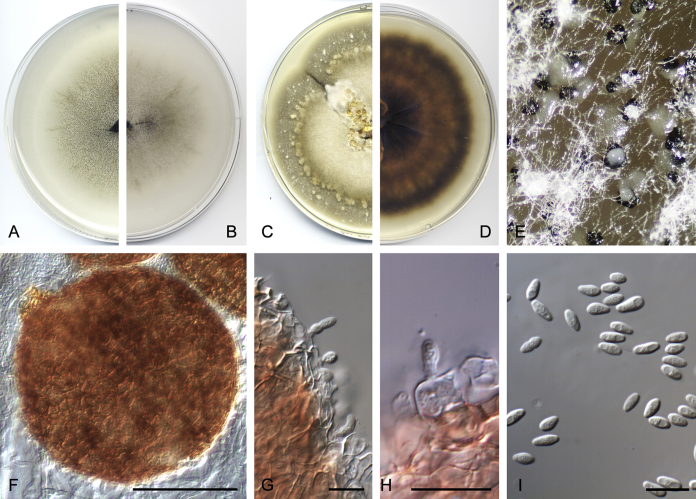
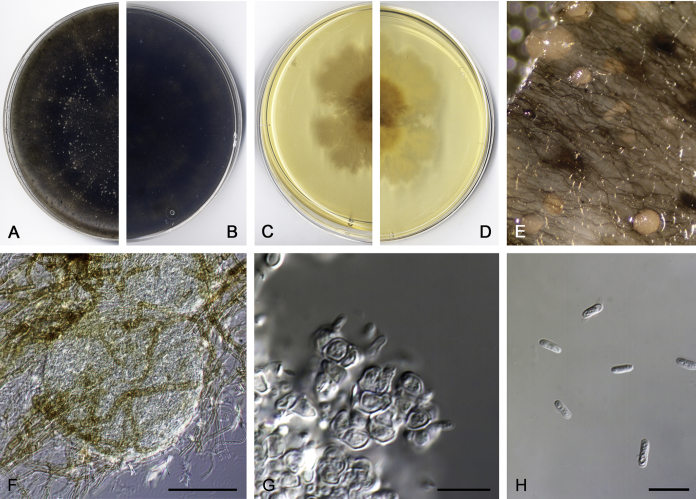
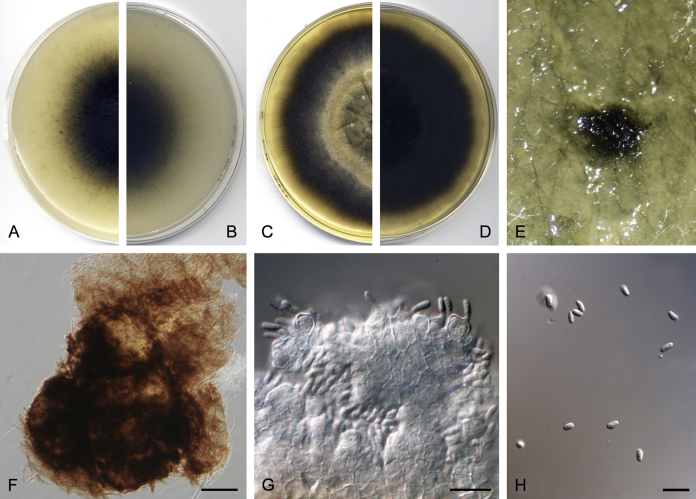
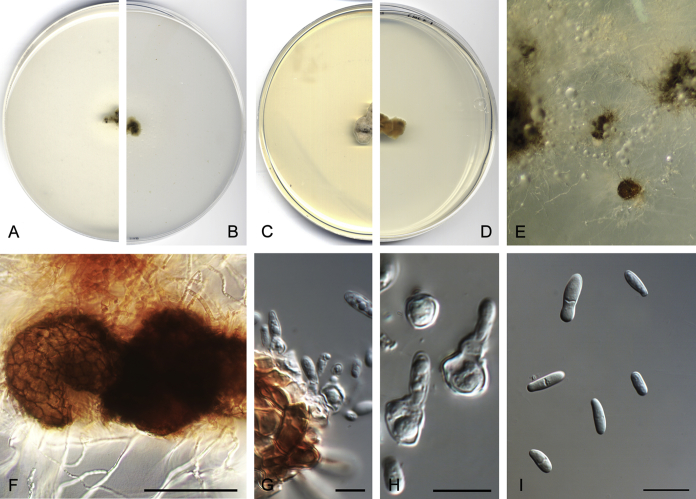
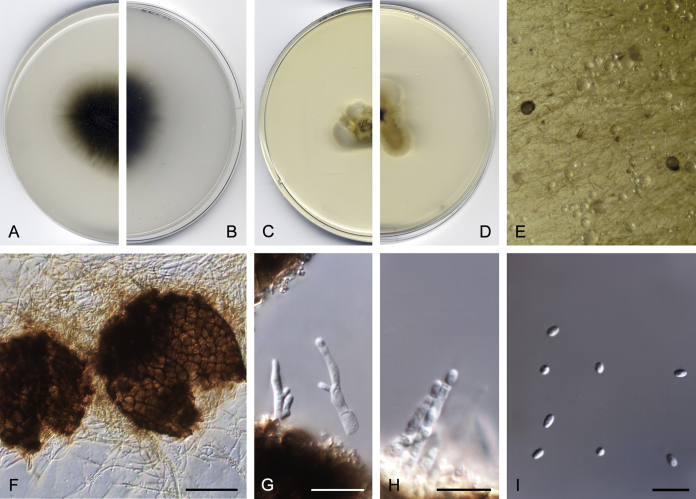
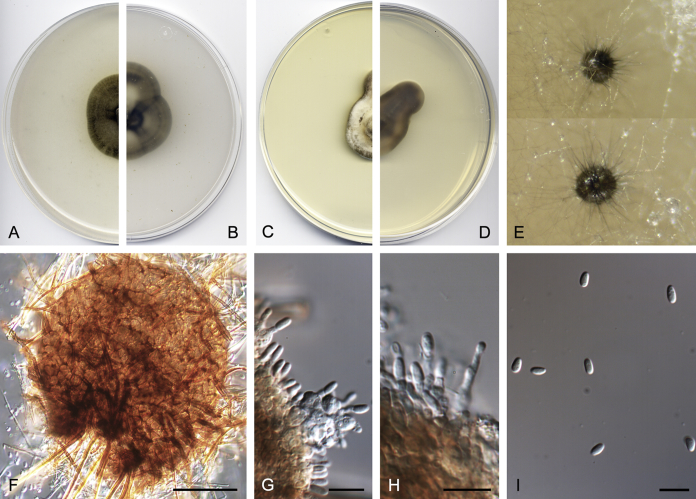
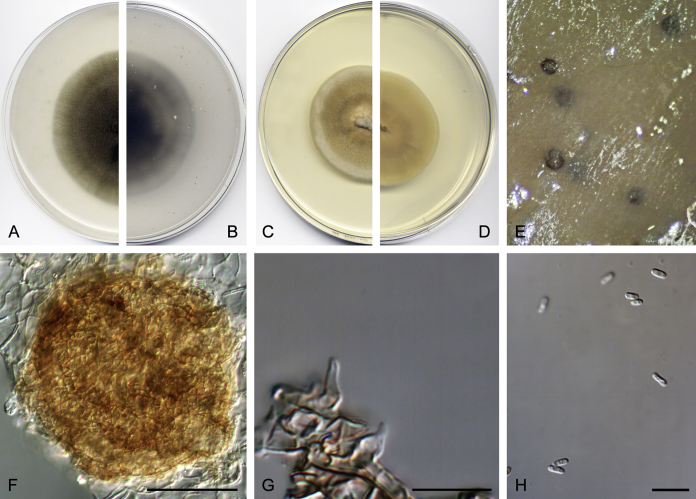
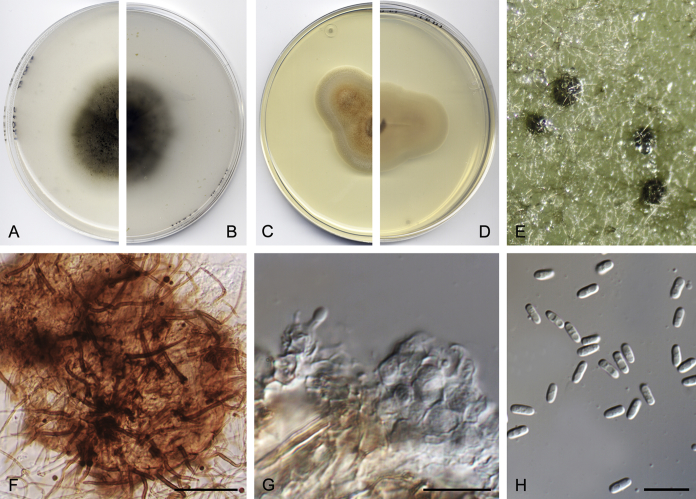
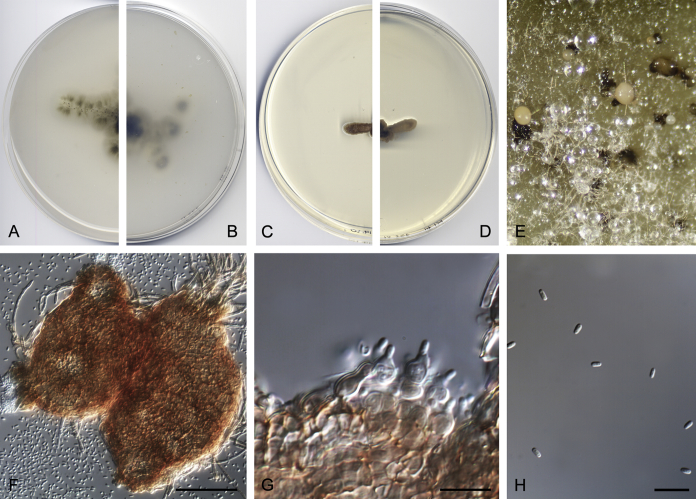
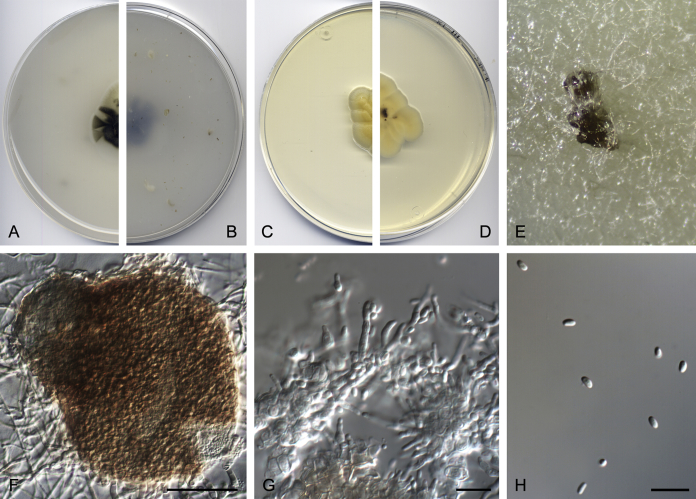
Didymella brunneospora Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, sp. nov. MycoBank MB820815. Fig. 2.
Fig. 2.
Didymella brunneospora (CBS 115.58). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin brunneus-, brown, and -spora, spore, because of the conidial pigmentation.
Description: Hyphae hyaline to pale brown, smooth- and thin-walled, septate, 2–5 μm wide. Conidiomata pycnidial, pale brown to dark brown, mostly solitary, occasionally confluent, superficial on OA, glabrous, globose, 140–250 μm diam, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–5 layered, 25–70 μm thick, composed of pale brown to brown, flattened polygonal cells of 8–15 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 7–10 × 6.5–8 μm. Conidia aseptate, hyaline to pale brown, smooth- and thin-walled, obovoid to cylindrical, 4.5–7 × 3–3.5 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 26 mm diam after 7 d at 25 ± 1 °C, flattened, with abundant production of pycnidia, olive brown (M. 4E6); reverse yellowish brown (M. 5E4). Colonies on MEA reaching 28 mm diam after 7 d at 25 ± 1 °C, flattened, orange melon (M. 5A6) to orange-white (M. 5A2); reverse orange melon (M. 5A6) to orange white (M. 5A2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: Germany, isolated from flower-stalk of Chrysanthemum roseum, R. Schneider (holotype CBS H-23199, ex-holotype living cultures IMB 8675 = DSM 62044 = CBS 115.58 = FMR 15745).
Notes: Ascochyta pyrethri (Brunaud 1887), reported on decaying stems of Pyrethri sinensis in Saintes (France), was originally described (very briefly and lacking of measurements of their reproductive structures) in French, but later Latinised by Saccardo (Saccardo 1892), changing the order of the authors. The description of that fungus by Saccardo was based on the original diagnosis: pycnidia conical-globose, sparse to arrange in linear series, erumpent, black; conidia numerous, ovoid, ellipsoidal or long ellipsoidal, somewhat obtuse at both ends, straight or slightly curved, subhyaline. However, Saccardo described the conidia as not being constricted at the septum, which was not mentioned in the original description. Moreover, the protologue lacks illustrations and references to herbarium material, which makes this taxon doubtful. The strain CBS 115.58, previously identified as Ascochyta pyrethri; clusters distant from Ascochyta and produces pale brown, aseptate conidia, features not seen in that genus, and thus being considered herein as a new species of the genus Didymella.
Didymella gardeniae (S. Chandra & Tandon) Q. Chen & L. Cai, Stud. Mycol. 82: 176. 2015.
Basionym: Pyrenochaeta gardeniae S. Chandra & Tandon, Mycopathol. Mycol. Appl. 29: 274. 1966.
Synonyms: Phoma gardeniae (S. Chandra & Tandon) Boerema, Verslagen Meded. Plantenziektenk. Dienst Wageningen 156: 27. 1980.
Peyronellaea gardeniae (S. Chandra & Tandon) Aveskamp et al., Stud. Mycol. 65: 32. 2010.
Description: de Gruyter & Boerema (2002).
Materials examined: India, Allahabad, from the leaf of Gardenia jasminoides, 1966, S. Chandra & R.N. Tandon (isotype CBS H-7605, ex-isotype living cultures CBS 626.68 = IMI 108771 = FMR 14901). USA, from human nail distrophy, 2006, D.A. Sutton, living cultures UTHSC DI16-211 = FMR 13701; from human toe nail, 2007, D.A. Sutton, living cultures UTHSC DI16-226 = FMR 13716; from human toe nails, 2009, D.A. Sutton, living cultures UTHSC DI16-274 = FMR 13765; from human wound neck, 2010, D.A. Sutton, living cultures UTHSC DI16-295 = FMR 13788.
Notes: Didymella gardeniae was first isolated from a leaf of Gardenia jasminoides in India (Chandra & Tandon, 1966), and it seems to be a common soil- and air-borne fungus recovered also from Netherlands Antilles. Here, it is for first time associated with human clinical specimens from North America. Morphologically our strains resemble D. gardeniae, but have setose pycnidia, which are more characteristic of Pyrenochaeta than phoma-like taxa. Also remarkable is the fact that our strains are capable of growing at 37 °C.
Didymella glomerata (Corda) Q. Chen & L. Cai, Stud. Mycol. 82: 176. 2015. Fig. 3.
Fig. 3.
Didymella glomerata (UTHSC DI16-205). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Alternaroid chlamydospores. H. Conidiogenous cells. I. Conidia. Scale bars: F = 100 μm. G–I = 10 μm.
Basionym: Coniothyrium glomeratum Corda, Icon. Fung. (Prague) 4: 39. 1840.
Synonyms: Phoma glomerata (Corda) Wollenw. & Hochapfel, Z. Parasitenk. 3: 592. 1936.
Peyronellaea glomerata (Corda) Goid. ex Togliani, Ann. Sperim. Agrar. III 6: 93. 1952.
Description: Boerema et al. (2004).
Materials examined: Lectotype designated here (MBT 377971): plate 8, fig. 108, in Corda, AKJ. 1840. Icones fungorum hucusque cognitorum. Tomus IV, Praga (http://bibdigital.rjb.csic.es/ing/Libro.php?Libro=1812). The Netherlands, from Chrysanthemum sp., 1963 (epitype designated here CBS H-16351, MBT377905, ex-epitype living cultures CBS 528.66 = PD 63/590). USA, from human superficial tissue sample, 2006, D.A. Sutton, living culture UTHSC DI16-205 = FMR 13695.
Notes: Coniothyrium glomeratum was introduced by Corda (1840). The description of this fungus is brief, and the illustrations are not very detailed. The natural source has been mentioned as dry greyed wood chips, but without any geographic location. No original material of the basionym exists. Therefore, we designate the illustration by Corda here as lectotype and CBS H-16351 as epitype of Coniothyrium glomeratum. Other authors placed this fungus in other genera, such as Aphosphaeria, Ascochyta, Peyronella and Phoma, but also in Alternaria, because the production of alternarioid chains of chlamydospores in vitro. For a complete discussion about synonymies of this fungus see Boerema et al. (1965), who gave an exhaustive morphological description in vitro of this fungus. Didymella glomerata is characterised by the production of subhyaline to carbonaceous, small to large, glabrous pycnidia bearing one (to two or three) ostioles, aseptate, hyaline to dark-coloured, ovoid to ellipsoidal conidia measuring mostly 6–7.5 × 3–3.5 μm, and alternaroid chlamydospores in chains. The fungus is distributed worldwide, and has been recovered from soil, different kinds of living and dead plants, and inorganic materials, and it can also infect humans (Punithalingam 1979, de Hoog et al. 2011). The strain UTHSC DI16-205, that phylogenetically clusters with the reference strain CBS 528.66 of Didymella glomerata, is morphologically indistinguishable from it.
Didymella heteroderae (Sen. Y. Chen et al.) Q. Chen & L. Cai, Stud. Mycol. 82: 176. 2015.
Basionym: Phoma heteroderae Sen Y. Chen et al., Mycologia 88: 885. 1996 (1997).
Synonyms: Peyronellaea heteroderae (Sen Y. Chen et al.) Crous, Persoonia 32: 223. 2014.
Phoma pomorum var. calorpreferens Boerema et al., Persoonia 15: 207. 1993.
Phoma calorpreferens (Boerema et al.) Aveskamp et al., Mycologia 101: 370. 2009.
Peyronellaea calorpreferens (Boerema et al.) Aveskamp et al., Stud. Mycol. 65: 31. 2010.
Description: Boerema (1993).
Materials examined: The Netherlands, from undefined food material, 1973, G.H. Boerema (holotype L 990.290.418, ex-holotype living cultures CBS 109.92 = PD 73/1405). USA, from human left plantar foot, 2005, D.A. Sutton, living cultures UTHSC DI16-190 = FMR 13680; from human nail, 2007, D.A. Sutton, living cultures UTHSC DI16-224 = FMR 13714; from human nail, 2007, D.A. Sutton, living cultures UTHSC DI16-227 = FMR 13717; from human fingernail, 2007, D.A. Sutton, living cultures UTHSC DI16-231 = FMR 13721; from human urine, 2007, D.A. Sutton, living cultures UTHSC DI16-232 = FMR 13722; from human nail, 2007, D.A. Sutton, living cultures UTHSC DI16-234 = FMR 13724; from human scalp, 2007, D.A. Sutton, living cultures UTHSC DI16-235 = FMR 13725; from human sputum sample, 2011, D.A. Sutton, living culturse UTHSC DI16-305 = FMR 13798.
Notes: Our strains are morphologically similar to the ex-type strain of D. heteroderae, and also show an identical DNA nucleotide sequence dataset. However, we proved that our strains are able to grow and sporulate at 37 °C (Valenzuela-Lopez et al. 2016), a higher temperature than that given in the original species description (Boerema et al. 2004).
Didymella keratinophila Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB820813. Fig. 4.
Fig. 4.
Didymella keratinophila (CBS 143032). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Section of pycnidium. H. Conidiogenous cells. I. Conidia. Scale bars: F, G = 100 μm. H, I = 10 μm.
Etymology: From Greg κɛρατινων-, keratin, and – φίλος, friend of, because the source from which the fungus was isolated.
Description: Hyphae pale brown, smooth- and thin-walled, septate, 2.5–8 μm wide. Conidiomata pycnidial, brown, solitary, superficial on OA, glabrous, broadly ellipsoidal, 250–270 × 200–230 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–5 layered, 15–35 μm thick, composed of brown, flattened polygonal cells of 5–10 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform or globose, 4.5–6 × 3–4.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, guttulate, ovoid to cylindrical, 4–6 × 2.5–3 μm. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 54 mm diam after 7 d at 25 ± 1 °C, flattened, greyish brown (M. 5F3); reverse greyish brown (M. 5F3). Colonies on MEA reaching 57–67 mm after 7 d at 25 ± 1 °C, flattened, brownish orange (M. 5C3); reverse brownish grey (M. 5C2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 37 °C.
Materials examined: USA, from human finger-hand lesion, 2006, D.A. Sutton (holotype CBS H-23200, ex-type living cultures CBS 143032 = UTHSC DI16-200 = FMR 13690); from human toe nail, 2007, D.A. Sutton, living cultures UTHSC DI16-228 = FMR 13718; from human nail, 2009, D.A. Sutton, living cultures UTHSC DI16-282 = FMR 13774.
Notes: Didymella keratinophila was recovered from a human superficial tissue specimen in the USA, and forms a well-supported sister clade with D. sancta. Didymella keratinophila differs phenotypically from D. sancta (and related species, such as D. glomerata, D. musae and D. pomorum) by the absence of chlamydospores in vitro (brown, alternarioid, phragmosporous and dyctiosporous, singly and terminally produced in D. sancta), smaller conidia (4–6 × 2.5–3 μm vs. 5–7 (–7.5) × 2.5–4 (–4.5) μm in D. sancta) and a negative NaOH spot test.
Didymella microchlamydospora (Aveskamp & Verkley) Q. Chen & L. Cai, Stud. Mycol. 82: 178. 2015.
Basionym: Phoma microchlamydospora Aveskamp & Verkley, Mycologia 101: 374. 2009.
Description: Aveskamp et al. (2009).
Materials examined: UK, from leaves of Eucalyptus sp., 1994, A.M. Ainsworth (holotype CBS H-20147, ex-holotype living culture CBS 105.95). USA, from human skin leg, 2006, D.A. Sutton, living cultures UTHSC DI16-199 = FMR 13689; from human corneal lesion, 2014, D.A. Sutton, living cultures UTHSC DI16-365 = FMR 13858.
Notes: Phoma microchlamydospora was described as a new species by Aveskamp et al. (2009) from leaves of Eucalyptus sp. in Great Britain, being subsequently transferred to the genus Didymella by Chen et al. (2015) after a phylogenetic study. Our two strains of this species differ in the geographic origin (USA) and in substrate (isolated from human clinical specimens), but they are morphologically and genetically similar to the ex-type living culture of D. microchlamydospora, being characterised by the production of abundant micropycnidia, globose pycnidia with 1–3 papillate ostioles, frequently on a neck, hyaline, one-celled, globose to ellipsoidal conidia, and relatively small, one-celled to multi-celled chlamydospores arranged in chains.
Didymella musae (P. Joly) Q. Chen & L. Cai, Stud. Mycol. 82: 178. 2015. Fig. 5.
Fig. 5.
Didymella musae (CBS 463.69). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Peyronellaea musae P. Joly, Rev. Mycol. 26: 97. 1961.
Synonym: Phoma jolyana Piroz. & Morgan-Jones, Trans. Brit. Mycol. Soc. 51: 200. 1968.
Description: Boerema (1993).
Materials examined: India, from fruit of Mangifera indica, May 1969, living cultures CBS 463.69 = FMR 15339. USA, from human cornea lesion, 2007, D.A. Sutton, living cultures UTHSC DI16-230 = FMR 13720.
Notes: The strain UTHSC DI16-230, which is morphologically similar to the reference strain CBS 463.69, only differs genetically in a few nucleotides of the tub2 gene.
Didymella protuberans (Lév.) Q. Chen & L. Cai, Stud. Mycol. 82: 180. 2015.
Basionym: Phoma protuberans Lév., Ann. Sci. Nat. Bot. III 5: 281. 1846.
Synonyms: Peyronellaea protuberans (Lév.) Aveskamp et al., Stud. Mycol. 65: 33. 2010.
Didymella alectorolophi Rehm, Hedwigia 64: 294. 1923.
Peyronellaea alectorolophi (Rehm.) Aveskamp et al., Stud. Mycol. 65: 31. 2010.
Phoma alecotorolophi Boerema et al., Persoonia 16: 366. 1997.
Phoma obtusa Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 378. 1870.
Peyronellaea obtusa (Fuckel) Aveskamp et al., Stud. Mycol. 65: 33. 2010.
Description: Chen et al. (2015).
Materials examined: The Netherlands, from a leaf of Lycium halimifolium, 1971 (neotype HMAS 246694, ex-neotype living cultures CBS 381.96 = PD 71/706). USA, from chocolate, 2011, D.A. Sutton, living cultures UTHSC DI16-302 = FMR 13795.
Notes: The strain UTHSC DI16-302 isolated from the USA clusters with the ex-neotype strain of Didymella protuberans, being morphologically similar.
Didymella rumicicola (Boerema & Loer.) Q. Chen & L. Cai, Stud. Mycol. 82: 181. 2015.
Basionym: Phoma rumicicola Boerema & Loer., New Zealand J. Bot. 18: 473. 1980.
Description: Chen et al. (2015).
Materials examined: New Zealand, Levin, from Rumex obtusifolius, Jun. 1979, G.F. Laundon (holotype PDD 50667, isotype CBS H-7627, ex-isotype living cultures CBS 683.79 = LEV 15094). The Netherlands, Baarn, from a stem of Rumex hydrolapathum, Mar. 1996, H.A. van der Aa, living culture CBS 179.97.
Notes: The strain CBS 179.97 was initially identified as Didymella acetosellae; this strain is genetically identical to the ex-type strain of D. rumicicola (CBS 683.79), and the host pertains to the same genus of plants (Rumex). Consequently, we assigned this strain to D. rumicicola.
Didymella sp.
Material examined: Japan, from Camellia sasanqua, living culture LC 8141.
Notes: This strain was considered by Chen et al. (2017) as a reference strain of Didymella segeticola. However, in our phylogenetic study, this strain is distinct from the ex-type strain of Didymella segeticola, with strain LC 8141 differing in 7 bp in rpb2. It was also isolated from a different host and country, and therefore we maintain this strain as Didymella sp.
Clade A2: Epicoccum
Epicoccum Link, Mag. Neuesten Entdeck. Gesammten Naturk. Ges. Naturf. Freunde Berlin 7: 32. 1815, emend. Q. Chen & L. Cai, Stud. Mycol. 82: 171. 2015.
Type species: Epicoccum nigrum Link.
Epicoccum camelliae Q. Chen et al., Stud. Mycol. 87: 140. 2017.
Description: Chen et al. (2017).
Materials examined: China, Jiangxi, Ganzhou, leaves of Camellia sinensis, 7 Sep. 2013, Y. Zhang (holotype HMAS 247159, ex-holotype culture CGMCC 3.18343 = LC 4858); ibid. LC4862. USA, from human respiratory tract, 2006, D.A. Sutton, living cultures UTHSC DI16-201 = FMR 13691; from human nail, 2006, D.A. Sutton, living cultures UTHSC DI16-202 = FMR 13692; from human toe nail, 2006, D.A. Sutton, living cultures UTHSC DI16-206 = FMR 13696; from human toe nail, 2009, D.A. Sutton, living cultures UTHSC DI16-280 = FMR 13772; from human nail, 2011, D.A. Sutton, living cultures UTHSC DI16-338 = FMR 13831; from human abscess, 2012, D.A. Sutton, living cultures UTHSC DI16-345 = FMR 13838.
Notes: A total of six isolates molecularly identified as E. camelliae clustered together with E. viticis forming a low-supported clade. Our isolates, as well as those of Chen et al. (2017), remained sterile. Consequently, further studies will be needed to fully characterise this species.
Epicoccum catenisporum Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, sp. nov., MycoBank MB819762. Fig. 6.
Fig. 6.
Epicoccum catenisporum (CBS 181.80). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Chlamydospores in chains. H. Conidiogenous cells. I. Conidia. Scale bars: F = 100 μm. G–I = 10 μm.
Etymology: From Latin catena-, chain, and -spora, spore, because of the disposition of the chlamydospores in chains.
Description: Hyphae pale brown, smooth- and thin-walled, septate, 2.5–5 μm wide. Conidiomata pycnidial, brown to dark brown, solitary, superficial and immersed (OA), glabrous, subglobose, 170–190 × 140–160 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–5 layered, 15–50 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–10 μm diam, Conidiogenous cells phialidic, hyaline, smooth-walled, doliiform or ampulliform, 4–6 × 2–4 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid or ellipsoidal, 4–5 × 2–3 μm, guttulate. Chlamydospores aseptate, dark brown, smooth- and thick-walled, in chains or singly, then intercalary disposed, ellipsoidal to ovoid, 9.5–12 × 4.5–8.5 μm.
Culture characteristics: Colonies on OA reaching 53 mm diam after 7 d at 25 ± 1 °C, flattened, powdery due to the production of abundant pycnidia, orange grey (M. 5B1) to yellowish brown (M. 5F5); reverse pale brown (M. 5D4) to brownish grey (M. 5F2). Colonies on MEA reaching 36 mm after 7 d at 25 ± 1 °C, flattened to floccose, white (M. 5A1) to orange white (M. 5A2); reverse white (M. 5A1) to pale orange (M. 5A4). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 35 °C.
Material examined: Guinea-Bissau, Gacheu, from a leaf spot of Oryza sativa, Oct. 1978, deposited by G.H. Boerema (holotype CBS H-23203, ex-holotype living cultures CBS 181.80 = PD 78/974 = FMR 14911).
Notes: The strain CBS 181.80 was previously identified as Phoma sorghina (currently E. sorghinum) by Aveskamp et al. (2009). However, it is phylogenetically different from that species. Epicoccum catenisporum is morphologically characterised by the production of pycnidia as observed in several other members of Epicoccum.
Epicoccum keratinophilum Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov., MycoBank MB819758. Fig. 7.
Fig. 7.
Epicoccum keratinophilum (CBS 142455). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cell. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Greek κɛράτινη-, keratin, and -φίλος, friend, linked to the origin of the fungus.
Description: Hyphae pale brown, smooth- and thin-walled, septate, 2.5–5 μm wide. Conidiomata pycnidial, brown, solitary, superficial or immersed (OA), glabrous, subglobose, 200–300 × 180–240 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–4 layered, 15–45 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–20 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform to globose, 4–5 μm diam. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 4–6 × 1.5–2 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 30–35 mm diam after 7 d at 25 ± 1 °C, flattened, entire edge, yellowish brown (M. 5E7) to brownish grey (M. 5F2); reverse brownish grey (M.5F2). Colonies on MEA reaching 30–37 mm diam after 7 d at 25 ± 1 °C, flattened, entire edge, white (M. 2A1) to yellowish white (M. 3A2); reverse dull yellow (M. 3B3). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Materials examined: USA, Texas, from animal skin lesion, 2009, D.A. Sutton (holotype CBS H-23032, ex-holotype living cultures CBS 142455 = UTHSC DI16-271 = FMR 13762); Texas, from human superficial tissue, 2007, D.A. Sutton, living cultures UTHSC DI16-244 = FMR 13734; from human bronchial wash sample, 2008, D.A. Sutton, living cultures UTHSC DI16-258 = FMR 13748; from human toe nail, 2009, D.A. Sutton, living cultures UTHSC DI16-272 = FMR 13763; from human biopsy tissue, 2011, D.A. Sutton, living culture UTHSC DI16-299 = FMR 13792.
Notes: In our phylogenetic tree E. keratinophilum forms a well-supported clade distant from its morphological relatives E. brasiliense and E. draconis. All E. keratinophilum strains have been recovered from clinical samples, and morphologically differ from E. brasiliense in producing smaller pycnidia and conidia, and from both E. brasiliense and E. draconis by a negative NaOH spot test reaction.
Epicoccum latusicollum Q. Chen et al., Stud. Mycol. 87: 144. 2017.
Description: Chen et al. (2017).
Materials examined: China, Jiangxi, Ganzhou, endophyte of Camellia sinensis, 7 Sep. 2013, Y. Zhang, living culture LC 4859; Shandong, Jining, on leaves of Sorghum bicolor, 3 Aug. 2013, N. Zhou (holotype HMAS 247164, ex-holotype living culture CGMCC 3.18346 = LC 5158). USA, from human eye, 2005, D.A. Sutton, living cultures UTHSC DI16-197 = FMR 13687.
Notes: The strain UTHSC DI16-197 that was isolated from a human eye sample clustered with the ex-type strain of E. latusicollum that was recently introduced by Chen et al. (2017), being characterised by the production of pycnidial conidiomata. Unfortunately, our strain was sterile, and morphological comparison was not possible, but genetically it is identical to the latter species.
Epicoccum ovisporum Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, sp. nov., MycoBank MB819761. Fig. 8.
Fig. 8.
Epicoccum ovisporum (CBS 180.80). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G, H. Conidiogenous cells. I. Conidia. Scale bars: F = 100 μm. G–I = 10 μm.
Etymology: From Latin ovum-, egg, and -spora, spore, due to the shape of the conidia.
Description: Hyphae hyaline to pale brown, smooth- and thin-walled, septate, 2.5–5 μm wide. Conidiomata pycnidial, brown, solitary, mostly superficial on OA and immersed into MEA, glabrous, subglobose to globose, 100–190 × 85–180 μm, with short papillate ostiolar neck; pycnidial wall of textura angularis, 3–4 layered, 12.5–35 μm thick, composed of brown, flattened polygonal cells of 5–20 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, doliiform to ampulliform, 5–6 × 2–3 μm. Conidia aseptate, hyaline, smooth- and thin-walled, guttulate, ovoid, ellipsoidal to cylindrical, 5–7 × 2–3 μm. Chlamydospores multi-celled, brown to dark brown, smooth-walled, disposed in chains or singly, then intercalary and terminally, globose to subglobose, 10–22.5 × 10–20 μm.
Culture characteristics: Colonies on OA reaching 36 mm diam after 7 d at 25 ± 1 °C, flattened, with abundant production of pycnidia, greenish grey (M. 29B2); reverse orange grey (M.5B2). Colonies on MEA reaching 38 mm after 7 d at 25 ± 1 °C, floccose, orange grey (M. 5B2) to grey (M. 5C1), producing a hyaline exudate; reverse yellowish brown (M. 5D8). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 30 °C.
Material examined: South Africa, Potchefstroom, from a leaf of Zea mays, Nov. 1978, isolated by W.J. Jooste, deposited by G.H. Boerema (holotype CBS H-23204, ex-holotype living cultures CBS 180.80 = PD 78/1100 = FMR 14910).
Notes: The strain CBS 180.80 was previously assigned to E. sorghinum (Aveskamp et al., 2009, Aveskamp et al., 2010); however, in our phylogenetic tree it represents a new species, forming a basal clade together with E. catenisporum and E. sorghinum, being distant from the rest of the species of the genus. The above-mentioned species are morphologically similar to E. ovisporum by producing pycnidia instead of sporodochia.
Epicoccum pneumoniae Valenzuela-Lopez, Stchigel, Guarro & Cano, sp. nov. MycoBank MB822112.
Etymology: The species name refers to the infection associated with this specimen.
Culture sterile. Epicoccum pneumoniae differs from its closest phylogenetic species Epicoccum latusicollum based on alignment of the concatenated four loci deposited in TreeBASE (S21115): LSU deletion in position: 382; ITS positions: 587 (C); tub2 positions: 1075 (T), 1102 (T), 1152 (T), 1159 (T), 1161 (G), 1207 (G), 1209 (T), 1210 (A), 1212 (G), 1213 (T), 1254 (T), 1260 (C), 1284 (C); rpb2 positions: 1312 (A), 1336 (A), 1339 (G), 1351 (C), 1354 (T), 1384 (C), 1453 (T), 1456 (C), 1495 (T), 1553 (C), 1609 (T), 1757 (T), 1769 (C), 1813 (C), 1816 (C), 1843 (C), 1873 (C), 1897 (C).
Culture characteristics: Colonies on OA reaching 29 mm diam after 7 d at 25 ± 1 °C, flattened, reddish grey (M. 9B2) to white (M. 9A1); reverse white (M. 9A1). Colonies on MEA reaching 31 mm after 7 d at 25 ± 1 °C, flattened to floccose, pinkish white (M. 9A2) to white (M. 9A1); reverse white (M. 9A1). NaOH spot test negative. Crystals absent.
Material examined: USA, from human sputum sample, 2008, D.A. Sutton (holotype FMR H-13747, ex-holotype living cultures UTHSC DI16-257 = FMR 13747).
Notes: The strain UTHSC DI16-257, which remained sterile, forms a basal clade with E. latusicollum; however, this strain clearly differs phylogenetically from the latter species mainly in the loci tub2 and rpb2. Therefore it is proposed here as a new species, Epicoccum pneumoniae.
Epicoccum proteae (Crous) Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, comb. nov. MycoBank MB820830.
Basionym: Phoma proteae Crous, Persoonia 27: 151. 2011.
Description: Crous et al. (2011).
Material examined: South Africa, Western Cape Province, Somerset West, Karibia Farm, from leaves of Protea cv. Carnival (P. compacta × P. neriifolia), 21 July 1998, J.E. Taylor & S. Denman (holotype CBS H-20771, ex-holotype living cultures CPC 1854 = CBS 114179 = FMR 15332).
Notes: This species was first proposed by Crous et al. (2011) within Phoma, which is characterised by producing brown, globose pycnidia and hyaline, aseptate conidia. However, our phylogenetic study showed that the ex-type strain of this species clustered in Epicoccum. Therefore, we propose a new combination for this species.
Epicoccum sorghinum (Sacc.) Aveskamp et al., Stud. Mycol. 65: 36. 2010.
Basionym: Phyllosticta sorghina Sacc., Michelia 1: 140. 1878.
Synonym: Phoma sorghina (Sacc.) Boerema et al., Persoonia 7: 134. 1973.
Description: Boerema et al. (2004).
Materials examined: France, Antibes, from a twig of Citrus sp., 1966, living cultures CBS 627.68 = PD 66/926. Puerto Rico, Mayaguez, from Sorghum vulgare, Apr. 1976, R. Alconera, living cultures CBS 179.80 = PD 76/1018; USA, from human foot, 2010, D.A. Sutton, living cultures UTHSC DI16-288 = FMR 13692; from human bronchial wash sample, 2011, D.A. Sutton, living cultures UTHSC DI16-301 = FMR 13794.
Notes: Two strains (UTHSC DI16-288 and UTHSC DI16-301) isolated from human clinical specimens in the USA clustered with the reference strains CBS 179.80 and CBS 627.68 of E. sorghinum. The latter species had been reported from several different substrates mainly from vegetal materials and it seems to be a widely distributed fungus, also having been associated with human infections (Punithalingam, 1985, Rai, 1989). Morphologically E. sorghinum was described producing mainly pycnidial conidiomata. Unfortunately, our strains were sterile, and further studies are needed to resolve the taxonomy of this species.
Clade A3: Allophoma
Allophoma Q. Chen & L. Cai, Stud. Mycol. 82: 162. 2015.
Type species: Allophoma tropica (R. Schneid. & Boerema) Q. Chen & L. Cai, Stud. Mycol. 82: 162. 2015.
Allophoma cylindrispora Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819625. Fig. 9.
Fig. 9.
Allophoma cylindrispora (CBS 142453). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidium. F. Conidiogenous cells. G. Conidia. Scale bars: E = 100 μm. F, G = 10 μm.
Etymology: From Latin cylindricus-, of cylindrical shape, and -spora, spore.
Description: Hyphae brown, septate, smooth- and thin-walled, 2.5–5 μm wide. Conidiomata pycnidial, brown to dark brown, confluent, superficial and immersed (OA), glabrous, ovoid, 120–210 × 90–140 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–4-layered, 15–30 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–12.5 μm diam,. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 3.5–4 × 4.5–5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 3–4 × 2 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 36 mm diam after 7 d at 25 ± 1 °C, flattened, beige (M. 4C3) to olive brown (M. 4F3); reverse blond (M. 4C4) to olive brown (M. 4F3). Colonies on MEA reaching 25–27 mm after 7 d at 25 ± 1 °C, flattened, white (M. 4A1); reverse pale yellow (M. 4A4) to yellowish orange (M. 4B7). NaOH spot test negative. Crystals absent. Optimal temperature of growth and of sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from a human eye lesion, 2007, D.A. Sutton (holotype CBS H-23030, ex-holotype living cultures CBS 142453 = UTHSC DI16-233 = FMR 13723).
Notes: This species forms a clade which is distinct from the closest relatives, A. minor and A. piperis. Unfortunately, the morphological distinction between these three species is difficult. Although these species differ in geography and substrate, molecular data is required for species identification. Allophoma cylindrispora sporulates poorly in culture.
Clade A7: Ectophoma
Ectophoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, gen. nov. MycoBank MB819952.
Etymology: From the Greek ɛκτοs, outside, because it is phylogenetically far from Phoma.
Conidiomata pycnidial, brown to dark brown, solitary or confluent, pycnidial wall of textura angularis, glabrous, globose to subglobose or irregular, ostiolate, with one or more short necks. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform to globose. Conidia aseptate, hyaline, smooth- and thin-walled, oblong to ellipsoidal, guttulate.
Type species: Ectophoma multirostrata (P.N. Mathur et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel.
Ectophoma multirostrata (P.N. Mathur et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, comb. nov., MycoBank MB819953. Fig. 10.
Fig. 10.
Ectophoma multirostrata (CBS 274.60). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Sphaeronaema multirostratum P.N. Mathur et al., Sydowia 13: 146. 1959.
Synonym: Phoma multirostrata (P.N. Mathur et al.) Dorenb. & Boerema, Mycopathol. Mycol. Appl. 50: 256. 1973.
Description: Boerema et al. (2004).
Materials examined: India, Maharashtra, Poona, Talegaon, from poultry farm soil, Mar. 1959, M. J. Thirumalachar (isotype CBS H-7616, ex-isotype living cultures CBS 274.60 = IMI 081598 = FMR 15335); Maharashtra, Poona, Talegaon, from soil, Mar. 1959, M.J. Thirumalachar, living cultures CBS 368.65 = PD 92/1757 = FMR 15336. The Netherlands, Hoorn, greenhouse, from the stem of Cucumis sativus, Aug. 1967, G.H. Boerema, living cultures CBS 110.79 = PD 65/8875 = FMR 15342.
Notes: Aveskamp et al. (2009) transferred this species from Sphaeronaema to Phoma. In our study, P. multirostrata forms a distinct clade, separated from all genera previously described in the Didymellaceae. Therefore, we propose a new genus to accommodate this species.
Ectophoma pomi (A.S. Horne) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, comb. nov. MycoBank MB819954. Fig. 11.
Fig. 11.
Ectophoma pomi (CBS 267.92). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Chlamydospores. H. Arrow indicate the conidiogenous cell. I. Conidia. Scale bars: F = 50 μm. G–I = 10 μm.
Basionym: Polyopeus pomi A.S. Horne, J. Bot., Lond. 58: 240. 1920.
Synonym: Phoma pereupyrena Gruyter et al., Persoonia 15: 398. 1993.
Description: Boerema et al. (2004).
Material examined: India, from a leaf spot of Coffea arabica, 1976, deposited by J. de Gruyter (neotype designated here CBS H-23202, MBT377913, ex-neotype living cultures CBS 267.92 = PD 76/1014 = FMR 15346).
Notes: Polyopeus pomi, introduced by Horne (1920) was validly described growing on potato mush agar, and was isolated from the fruits of Malus domestica “Cox's Orange Pippin”, in the UK, where it produced dark spots. No illustration is available, and no type material is mentioned in the publication. Therefore, based on the original description, we propose CBS H-23202 as neotype. The fungus produces black, subglobose to irregularly shaped pycnidia with a blackish neck, and hyaline, ellipsoidal conidia, 5−9 × 2−3 μm.
Clade A8: Remotididymella
Remotididymella Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, gen. nov. MycoBank MB819990.
Etymology: From Latin remotus-, distant, because it is phylogenetic far removed from the similar genus Didymella.
Conidiomata pycnidial, brown to dark brown, mostly confluent; pycnidial wall of textura angularis, mostly glabrous, globose or irregularly-shaped, with a single ostiole. Conidiogenous cells phialidic, hyaline, smooth-walled, globose or ampulliform. Conidia aseptate, hyaline, smooth- and thin-walled, allantoid or cylindrical, guttulate.
Type species: Remotididymella destructiva (Plowr.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel.
Remotididymella anthropophila Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819991. Fig. 12.
Fig. 12.
Remotididymella anthropophila (CBS 142462). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Greek ανθρώπος-, human, and –φίλος, friend, because that fungus has been isolated from a human sample.
Description: Hyphae brown, smooth- and thin-walled, septate, 2.5–8 μm wide. Conidiomata pycnidial, apricot to pale brown, translucent, solitary or confluent, superficial (OA), glabrous, globose to subglobose, 300–400 × 250–400 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–5 layered, 30–40 μm thick, composed of subhyaline to pale brown flattened polygonal cells of 5–20 μm diam,. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform to globose, 5–6 μm diam. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 5.5–7.5 × 1.5–2.5 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 60 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish brown (M. 5E3) to greyish brown (M. 5F3); reverse greyish brown (M. 5F3). Colonies on MEA reaching 35–36 mm diam after 7 d at 25 ± 1 °C, flattened, greyishorange (M. 5B3) to pale brown (M. 5D6); reverse orange white (M. 5A2) to brownish yellow (M. 5C7). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 35 °C.
Material examined: USA, Texas, from human bronchial secretion, D.A. Sutton (holotype CBS H-23039, ex-holotype living cultures CBS 142462 = UTHSC DI16-278 = FMR 13770).
Notes: The new species Remotididymella anthropophila is genetically distinct from its nearest neighbour R. destructiva. Morphologically it is the only species of the genus that produces pale-brown pycnidia, which is unusual in phoma-like species, and it differs in substrate and location with the latter species.
Remotididymella destructiva (Plowr.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, comb. nov. MycoBank MB819992. Fig. 13.
Fig. 13.
Remotididymella destructiva (CBS 378.73). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Phoma destructiva Plowr., Gard. Chron. II 16: 621. 1881.
Synonyms: Diplodina destructiva (Plowr.) Petr., Annls mycol. 19(1/2): 19. 1921.
Phoma destructiva var. diversispora Gruyter et al., Persoonia 18: 28. 2002.
Description from ex-epitype (CBS 378.73): Hyphae brown, smooth- and thin-walled, septate, 2.5–6 μm wide. Conidiomata pycnidial, dark brown, mostly confluent, rarely solitary, superficial or immersed (OA), glabrous, ovoid to irregularly-shaped, 120–250 × 90–180 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–4 layered, 12.5–50 μm thick, composed of brown, flattened polygonal cells of 5–10 μm diam,. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 10–12 × 5–6 μm. Conidia aseptate, hyaline, smooth- and thin-walled, variable in shape, mostly allantoid to cylindrical, 3.5–8 × 2–2.5 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 21 mm diam after 7 d at 25 ± 1 °C, flattened, front and reverse dark grey (M. 4F1). Colonies on MEA reaching 10 mm diam after 7 d at 25 ± 1 °C, flattened, front and reverse olive brown to dark grey (M. 5F2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 30 °C.
Materials examined: Lectotype designated here (MBT378116): fig. 123, in Plowright. 1881. The Gardeners' chronicle: a weekly illustrated journal of horticulture and allied subjects (http://www.biodiversitylibrary.org/item/84372#page/639/mode/1up). Guadeloupe, from fruit of Lycopersicon esculentum, 1987, living cultures CBS 133.93 = PD 88/961 = IMI 173142. The Netherlands, Berkel en Rodenrijs, from a leaf of Lycopersicon esculentum, Oct. 1977, G.H. Boerema, living culture CBS 162.78 = PD 77/725. Tonga, Friendly Islands, from decaying fruit of Lycopersicon esculentum, 1967, G.F. Laundon (epitype designated here CBS H-16200, MBT377914, ex-epitype living cultures CBS 378.73 = FMR 15328 = CECT 2877).
Notes: Phoma destructiva was originally described by Plowright (1881), infecting fruits of Lycopersicon esculentum in King’s Lynn, UK. Later, many representative specimens were collected from the similar hosts in other countries of Europe, and in North and South America (De Gruyter et al., 2002, Boerema et al., 2004). Phoma destructiva is characterised by the production of olivaceous black, globose, glabrous pycnidia with up to three papillate ostioles, hyaline, aseptate, subglobose to ellipsoidal conidia of σ = 5.8 × 2.2 μm, scarce and larger 1-septate conidia, and by the absence of chlamydospores. De Gruyter et al. (2002), based on morphological differences of the conidia, recognized two varieties, destructiva and diversispora. However, the isolates CBS 378.73 and CBS 133.93, representative strains of “Phoma destructiva var. destructiva”, and CBS 162.78, representative of “Phoma destructiva var. diversispora”, were phylogenetically and morphologically very similar in our study. Therefore, we did not accept these varieties, and propose CBS H-16200 as the epitype of Remotididymella destructiva.
Clade A9: Similiphoma
Similiphoma Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, gen. nov., MycoBank MB820847.
Etymology: From Latin similis-, similar to, due to the morphological similarity with Phoma.
Conidiomata pycnidial, brown, confluent or solitary; pycnidium wall of textura angularis, glabrous or with short hyphal outgrowths, globose to subglobose, with one or two papillate ostioles. Conidiogenous cells phialidic, hyaline, smooth-walled, globose or ampulliform. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, guttulate.
Type species: Similiphoma crystallifera (de Gruyter et al.) Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel.
Similiphoma crystallifera (Gruyter et al.) Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, comb. nov. MycoBank MB820848. Fig. 14.
Fig. 14.
Similiphoma crystallifera (CBS 193.82). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Phoma crystallifera Gruyter et al., Persoonia 15: 393. 1993.
Description: Boerema et al. (2004).
Material examined: Austria, Kärnten, Wallersberg near Völkermarkt, from Chamaespartium sagittale, 1982, H.A. van der Aa (holotype L 992.177-456, ex-holotype living cultures CBS 193.82 = FMR 15343).
Notes: Similiphoma crystallifera CBS 193.82 clustered phylogenetically distant from the closest morphologically related genera Ectophoma, Epicoccum and Phoma. Consequently, we designated this strain as the type species of the new genus Similiphoma.
Clade A10: Paraboeremia
Paraboeremia Q. Chen & L. Cai, Stud. Mycol. 82: 183. 2015.
Type species: Paraboeremia selaginellae (Sacc.) Q. Chen & L. Cai.
Paraboeremia putaminum (Speg.) Q. Chen & L. Cai, Stud. Mycol. 82: 184. 2015. Fig. 15.
Fig. 15.
Paraboeremia putaminum (CBS 130.69). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Phoma putaminum Speg., Atti Soc. Crittog. Ital. 3: 66. 1881.
Description: de Gruyter & Noordeloos (1992).
Material examined: Denmark, from the rhizosphere of Malus sylvestris, Mar. 1968, E. Sønderhousen, living cultures CBS 130.69 = CECT 20054 = IMI 331916 = FMR 15338.
Notes: This species was introduced by Spegazzini in 1881, isolated from pine wood in Sweden, and in the last study of this species by Chen et al. (2015) from two reference strains (CBS 130.69 and CBS 372.91) was placed within the genus Paraboeremia. However, without an illustration and rpb2 sequences, in our study, the rpb2 sequence and the illustration were provided of the reference strain CBS 130.69, which it resembles morphologically (de Gruyter & Noordeloos 1992), but further studies are needed to clarify its typification.
Paraboeremia selaginellae (Sacc.) Q. Chen & L. Cai, Stud. Mycol. 82: 184. 2015.
Basionym: Phyllosticta selaginellae Sacc., Malpighia 11: 304. 1897.
Synonym: Phoma selaginellicola Gruyter et al., Persoonia 15: 399. 1993.
Description: Chen et al. (2015).
Material examined: The Netherlands, from a leaf of Selaginella sp., 1977, G.H. Boerema (neotype HMAS 246693, MBT202501, ex-neotype living cultures CBS 122.93 = PD 77/1049 = FMR 15348).
Notes: This species was already typified by Chen et al. (2015) providing DNA sequence data and illustrations. However, the rpb2 sequence was not given, and therefore in the present study the rpb2 sequence of the ex-type strain CBS 122.93 is added.
Clade A12: Cumuliphoma
Cumuliphoma Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, gen. nov. MycoBank MB819878.
Etymology: From Latin cumulus-, heap or pile, in reference to the aggregated pycnidia.
Conidiomata pycnidial, brown, mostly confluent, pycnidial wall of textura angularis, mostly glabrous, globose or nearly so, with a single ostiole. Conidiogenous cells phialidic, hyaline, smooth-walled, globose to ampulliform. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, guttulate. Chlamydospores mostly absent.
Type species: Cumuliphoma omnivirens (Aveskamp et al.) Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano.
Cumuliphoma indica Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, sp. nov. MycoBank MB819880. Fig. 16.
Fig. 16.
Cumuliphoma indica (CBS 654.77). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: The name refers to the geographic origin of the fungus, India.
Description: Hyphae pale brown to brown, smooth- and thin-walled, septate, 2.5–8 μm wide. Conidiomata pycnidial, brown to dark brown, mostly confluent, rarely solitary, immersed (OA and MEA), glabrous, ovoid to irregularly-shaped, 150–180(–520) × 140–150(–490) μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 3–5-layered, 25–60 μm thick, composed of brown, flattened polygonal cells of 7–23 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, globose to ampulliform, 5–6 × 4–5.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, 4–5.5 × 2–2.5 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 42 mm diam after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4F3); reverse dark grey (M. 4F1). Colonies on MEA reaching 37 mm diam after 7 d at 25 ± 1 °C, flattened, brownish grey (M. 5F2) to pale grey (M. 5C2); reverse brownish grey (M. 5F2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 30 °C.
Materials examined: India, Jabalpur, from an unknown substrate, 1977, isolated by D.P. Tiwari (holotype CBS H-20152, ex-holotype living cultures CBS 654.77 = FMR 15341). Papua New Guinea, Varirata National Park, from soil, Aug. 1995, A. Aptroot, living cultures CBS 991.95 = FMR 15331.
Notes: The isolates CBS 654.77 and CBS 991.95 were received as “Phoma omnivirens”. However, these isolates were phylogenetically distant from the ex-type strain of C. omnivirens (CBS 341.86), and also both differ morphologically from the latter due to the absence of chlamydospores and micropycnidia.
Cumuliphoma omnivirens (Aveskamp et al.) Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, comb. nov. MycoBank MB819882.
Basionym: Phoma omnivirens Aveskamp et al., Mycologia 101: 375. 2009.
Description: Aveskamp et al. (2009).
Material examined: Belgium, Gembloux, from Phaseolus vulgaris, 1968, isolated by L. Obando (holotype CBS H-20151, ex-holotype living cultures CBS 341.86 = FMR 14915).
Notes: Cumuliphoma omnivirens is the only species of the genus that produces chlamydospores. Phylogenetically, it is closely related to C. pneumoniae, but is distinct from this species in both rpb2 and tub2 sequences by 9 bp.
Cumuliphoma pneumoniae Valenzuela-Lopez, Stchigel, Crous, Guarro & Cano, sp. nov. MycoBank MB819881. Fig. 17.
Fig. 17.
Cumuliphoma pneumoniae (CBS 142454). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Greek πνɛυμονικός-, pulmonary, due to the origin of the ex-type strain.
Description: Hyphae hyaline to brown, smooth- and thin-walled, septate, 2.5–6 μm wide. Conidiomata pycnidial, brown to dark brown, confluent, superficial (OA), glabrous, globose to subglobose, 200–240 × 200 μm, with a short papillate ostiolar neck; pycnidial wall of textura angularis, 3–5 layered, 25–35 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–12 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform to globose, 5–6 × 5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to cylindrical, 2.5–5 × 2 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 28 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish brown (M. 5F5); reverse brownish grey (M. 5F3). Colonies on MEA reaching 27–29 mm after 7 d at 25 ± 1 °C, flattened, grey (M. 6C1), producing a diffusible greyish orange pigment; reverse dark brown (M. 6F6). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human sputum sample, D.A. Sutton (holotype CBS H-23031, ex-holotype living cultures CBS 142454 = UTHSC DI16-249 = FMR 13739).
Notes: Cumuliphoma pneumoniae was isolated from a clinical sample of the respiratory tract. This species is morphologically closely related to C. omnivirens, which is also the phylogenetically nearest species. However, C. pneumoniae does not produce chlamydospores.
Clade A13: Juxtiphoma
Juxtiphoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, gen. nov. MycoBank MB821111.
Etymology: From Latin juxta, next to, due to the morphological and phylogenetic similarity with Phoma.
Conidiomata pycnidial, brown, mostly solitary, sometimes confluent, pycnidial wall of textura angularis, glabrous, subglobose to conical, papillate, ostiolate. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid, ellipsoidal or cylindrical, biguttulate. Chlamydospores aseptate, ochraceous-brown, single or in chains, subglobose, barrel-shaped or ellipsoidal.
Type species: Juxtiphoma eupyrena (Sacc.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel.
Juxtiphoma eupyrena (Sacc.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, comb. nov. MycoBank MB821112.
Basionym: Phoma eupyrena Sacc., Michelia 1: 525. 1879.
Description: Boerema et al. (2004).
Materials examined: Germany, Kiel-Kitzeberg, from wheat field soil, 1966, W. Gams, living cultures CBS 527.66 = FMR 15337 = ATCC 22238. The Netherlands, from the tuber of Solanum tuberosum, 1991, J. de Gruyter, living cultures CBS 374.91 = PD 78/391 = FMR 15329.
Notes: Phoma eupyrena, introduced by Saccardo (1879) and reported on stems of Solanum tuberosum (geographic origin not cited), has been revised by several authors. The description from Saccardo is minimal: blackish, depressed conical, ostiolate pycnidia with hyaline, ovoid conidia, 4 × 1.5 μm. Boerema et al. (2004) characterised this species morphologically and placed it in the section Phoma. Aveskamp et al. (2009) considered it phylogenetically close to “Phoma omnivirens”, and later Aveskamp et al. (2010) regarded P. eupyrena closely related to Microsphaeropsis. However, in our phylogenetic tree this species formed a well-supported monophyletic clade, separate from the other genera of Didymellaceae. Therefore, we propose the new genus Juxtiphoma to accommodate this species.
Clade A14: Vacuiphoma
Vacuiphoma Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, gen. nov. MycoBank MB821451.
Etymology: Based on the occurrence of empty pycnidial structures.
Conidiomata pycnidial, brown to dark brown, solitary, glabrous, subglobose or obpyriform; pycnidial wall of textura angularis, non-papillate.
Type species: Vacuiphoma bulgarica (Aveskamp et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel.
Vacuiphoma bulgarica (Aveskamp et al.) Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, comb. nov. MycoBank MB821452.
Basionym: Phoma bulgarica Aveskamp et al., Stud. Mycol. 65: 47. 2010.
Description: Aveskamp et al. (2010).
Material examined: Bulgaria, Silkossia, Strandga Mountain, from leafs of Trachystemon orientale, 20 Jun. 1980, S. Vanev (holotype CBS H-20242, ex-holotype living cultures CBS 357.84 = FMR 14917).
Notes: This species was introduced by Aveskamp et al. (2010) within the genus Phoma due to the production of pycnidial conidiomata. However, this species was not able to produce conidia and remains poorly characterised. Genetically this species along with V. oculihominis form a distinct clade within Didymellaceae, thus we treat these species within the new genus Vacuiphoma.
Vacuiphoma oculihominis Valenzuela-Lopez, Stchigel, Guarro & Cano, sp. nov. MycoBank MB822113.
Etymology: The epithet refers to the human eye clinical sample, from which the fungus was isolated.
Culture sterile. Vacuiphoma oculihominis differs from its closest phylogenetic species, Vacuiphoma bulgarica, in two bp of the ITS nucleotide sequence, 12 bp of tub2 and 44 bp of rpb2, based on alignment of the concatenated four loci deposited in TreeBASE (S21115).
Culture characteristics: Colonies on OA reaching 30–34 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish grey (M. 2B2) to olive grey (M. 2E2); reverse white (M. 2A1) to olive grey (M. 2E2). Colonies on MEA reaching 33 mm diam after 7 d at 25 ± 1 °C, slightly floccose, white (M. 5A1) to light orange (M. 5A4); reverse light orange (M. 5A4). NaOH spot test negative. Crystals absent. Optimal temperature of growth 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, Illinois, from human eye secretion, 2011, D.A. Sutton (holotype FMR H-13801, ex-holotype living cultures UTHSC DI16-308 = FMR 13801).
Notes: The strain UTHSC DI16-308 was recovered from a human eye clinical specimen, and remained sterile despite being cultured on different types of media. Because this strain is phylogenetically related with V. bulgarica, but distant from that species, it is proposed here as a new taxon.
Clade A15: Nothophoma
Nothophoma Q. Chen & L. Cai, Stud. Mycol. 82: 212. 2015.
Type species: Nothophoma infossa (Ellis & Everh.) Q. Chen & L. Cai, Stud. Mycol. 82: 213. 2015.
Nothophoma gossypiicola (Gruyter) Q. Chen & L. Cai, Stud. Mycol. 82: 213. 2015.
Basionym: Phoma gossypiicola Gruyter, Persoonia 18: 96. 2002.
Description: de Gruyter (2002).
Materials examined: USA, Texas, from a leaf of Gossypium sp., 1963, L.S. Bird, living cultures CBS 377.67 = FMR 14912; from human ethmoid sinus lesion, 2010, D.A. Sutton, living cultures UTHSC DI16-294 = FMR 13787.
Notes: This species was recently placed within the genus Nothophoma by Chen et al. (2015). In our study, one isolate from human clinical specimen was identified as N. gossypiicola, which it resembles in both morphology and DNA sequences from the reference strain CBS 377.67, isolated from the same country. This species is morphologically characterised by producing longer conidia (10–12.5 × 2.5–3.5 μm) and chlamydospores arranged in chains. However, further studies are needed to resolve its typification.
Nothophoma macrospora Valenzuela-Lopez et al., Persoonia 36: 431. 2016.
Description: Crous et al. (2016b).
Material examined: USA, Arizona, Phoenix, from human respiratory secretion of a patient with pneumonia, 1 Apr. 2009, D.A. Sutton (holotype CBS H-22377, ex-holotype living cultures CBS 140674 = UTHSC DI16-276 = FMR 13767).
Notes: This species was recently proposed by Valenzuela-Lopez et al. (2016), which is phylogenetically related with N. gossypiicola, but differs morphologically from the latter species in pycnidial shape, conidia (up to 2 vs non-septate) and the absence of chlamydospores (see Crous et al. 2016b). Furthermore, here the sequence of rpb2 is provided and differs in 13 bp from N. gossypiicola, and therefore N. macrospora is also phylogenetically distinct from N. gossypiicola.
Nothophoma quercina (Syd.) Q. Chen & L. Cai, Stud. Mycol. 82: 213. 2015.
Basionym: Cicinobolus quercinus Syd., Ann. Mycol. 13: 42. 1915.
Synonyms: Ampelomyces quercinus (Syd.) Rudakov, Mikol. Fitopatol. 13: 109. 1979.
Phoma fungicola Aveskamp et al., Stud. Mycol. 65: 26. 2010.
Description: Aveskamp et al. (2010).
Materials examined: Ukraine, Crimea, in the vicinity of Feodosiya, on Microsphaera alphitoides from Quercus sp., 1979, O.L. Rudakov living cultures CBS 633.92 = ATCC 36786, VKM MF-325 = FMR 14913. USA, from human superficial foot lesion, 2009, D.A. Sutton, living cultures UTHSC DI16-270 = FMR 13761.
Notes: This species was already accommodated by Chen et al. (2015) within Nothophoma, and is characterised by producing globose to suboblate, glabrous, solitary pycnidia and hyaline, aseptate conidia (see Aveskamp et al. 2010). In our study, one human clinical strain isolated in the USA clustered with the reference strain CBS 633.92 of N. quercina. Morphologically it resembles the latter strain, and only a few differences in bp were genetically noted. However, both strains form a well-supported clade and were identified as the same species.
Nothophoma variabilis Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819624. Fig. 18.
Fig. 18.
Nothophoma variabilis (CBS 142457). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Latin variabilis, due to the variable shape of the conidia.
Description: Hyphae pale brown, septate, smooth- and thin-walled, 2.5–6 μm wide. Conidiomata pycnidial, brown, confluent, superficial (OA), glabrous, subglobose, 150–350 × 130–270 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 3–6-layered, 25–35 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–20 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 6 × 5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical or irregularly shaped, 4–7 × 3–3.5 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 31 mm diam after 7 d at 25 ± 1 °C, flattened, greyish yellow (M. 4B4) to olive brown (M. 4F3); reverse olive brown (M. 4F3). Colonies on MEA reaching 36 mm after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4F3) to greyish yellow (M. 4C5); reverse olive brown (M. 4F3) to brownish grey (M. 4F2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and of sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 35 °C.
Material examined: USA, from human bronchial wash sample, 2009, D.A. Sutton (holotype CBS H-23034, ex-holotype living cultures CBS 142457 = UTHSC DI16-285 = FMR 13777).
Notes: This species was recovered from a clinical specimen of the respiratory tract, and it is closely related to N. anigozanthi. Both species can be differentiated by the presence of a single pycnidial ostiole (vs. 1–4 in N. anigozanthi), absence of a neck and production of wider conidia (3–3.5 μm vs. 1.5–2.5 μm) in N. variabilis. The NaOH spot test was negative, whereas it produces a dull green to vinaceous black pigmentation in N. anigozanthi.
Clade A21: Phoma
Phoma Sacc., Michelia 2: 4. 1880. emend. Q. Chen & L. Cai, Stud. Mycol. 82: 194. 2015.
Synonym: Atradidymella M.L. Davey & Currah, Amer. J. Bot. 96: 1283. 2009.
Type species: Phoma herbarum Westend.
Phoma herbarum Westend., Bull. Acad. Roy. Sci. Belgique, Cl. Sci. 19: 118. 1852. emend. Chen et al., Stud. Mycol. 82: 195. 2015.
Synonyms: Atradidymella muscivora M.L. Davey & Currah, Amer. J. Bot. 96: 1283. 2009.
Phoma muscivora M.L. Davey & Currah, Amer. J. Bot. 96: 1283. 2009.
Phoma cruris-hominis Punith., Nova Hedwigia 31: 135. 1979.
Description: Chen et al. (2015).
Materials examined: The Netherlands, Emmeloord, from the stem of Rosa multiflora cv. Cathayensis, Apr. 1965, G.H. Boerema, living cultures CBS 615.75 = PD 73/665 = IMI 199779 = FMR 15340; Naaldwijk, from a stem base of Nerium sp., 1986, J. de Gruyter, living cultures CBS 502.91 = PD 82/276. UK, from a leg of woman, Apr. 1977, Y.M. Clayton, holotype of “Phoma cruris-hominis” IMI 213845, living cultures CBS 377.92 = IMI 213845. USA, from human urine catheter, 2006, D.A. Sutton, living cultures UTHSC DI16-204 = FMR 13694; from human bronchial wash sample, 2006, D.A. Sutton, living cultures UTHSC DI16-212 = FMR 13702; from human sputum sample, 2011, D.A. Sutton, living cultures UTHSC DI16-306 = FMR 13799; from human bronchial sample, 2011, D.A. Sutton, living cultures UTHSC DI16-307 = FMR 13800; from human nail, 2010, D.A. Sutton, living cultures UTHSC DI16-319 = FMR 13812.
Notes: In this study five strains from human clinical specimens were identified as Phoma herbarum, all of them corresponding in morphology and genetically with the reference strains CBS 377.92, CBS 502.91 and CBS 615.75. This species was already described as an opportunistic human pathogenic fungus by Punithalingam (1979), and this fact is confirmed in our study.
Clade A24: Xenodidymella
Xenodidymella Q Chen et al., Stud. Mycol. 82: 205. 2015.
Type species: Xenodidymella applanata (Niessl) Q. Chen & L. Cai.
Xenodidymella saxea (Aveskamp et al.) Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, comb. nov. MycoBank MB820831.
Basionym: Phoma saxea Aveskamp et al., Stud. Mycol. 65: 23. 2010.
Description: Aveskamp et al. (2010).
Material examined: Germany, Oldenburg, from corroded Mediterranean marble, June 1992, J. Kuroczkin (holotype CBS H-20240, ex-holotype cultures CBS 419.92 = FMR 15347).
Notes: This species was introduced by Aveskamp et al. (2010) and placed together with “Phoma humicola” (currently Xenodidymella humicola). In our phylogenetic tree, this species was related to the Xenodidymella clade. Despite that this species could represent another genus based on its low phylogenetic support and morphology, more studies are needed to resolve its taxonomic placement in the Didymellaceae. Thus, a new combination is proposed for this species. Morphologically X. saxea is characterised by producing dimorphic conidia: I) aseptate, hyaline, smooth- and thin-walled, (sub-) globose, (3–)3.5–5.5 μm diam, guttulate; and II) aseptate, hyaline, smooth- and thin-walled, cylindrical to ellipsoidal, (3.5–)4.5–7(–7.5) × 2.5–3.5(–4) μm.
Clade A25: Neodidymelliopsis
Neodidymelliopsis Q. Chen et al., Stud. Mycol. 82: 207. 2015.
Type species: Neodidymelliopsis cannabis (G. Winter) Q. Chen & L. Cai.
Neodidymelliopsis longicolla L.W. Hou et al., Stud. Mycol. 87: 153. 2017.
Description: Chen et al. (2017).
Materials examined: Israel, En Avdat, Negev desert, from soil, Feb. 1996, A. van Iperen (holotype CBS H-23016, ex-holotype living culture CBS 382.96). USA, from human bronchial wash sample, 2011, D.A. Sutton, living cultures UTHSC DI16-322 = FMR 13815.
Notes: This species was recently proposed by Chen et al. (2017), and is characterised by producing globose to flask-shaped, glabrous or pycnidia with hyphal outgrowths. The most characteristic features include its elongated neck, the conidia that are initially hyaline and aseptate, but became pale-brown and septate with age. In our study, the strain UTHSC DI16-322 clustered with the ex-type strain of N. longicolla. However, no morphological comparison was possible because our strain remained sterile.
Clade A26: Neoascochyta
Neoascochyta Q. Chen& L. Cai, Stud. Mycol. 82: 198. 2015.
Type species: Neoascochyta exitialis (Morini) Q. Chen & L. Cai, Stud. Mycol. 82: 199. 2015.
Neoascochyta cylindrispora Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819691. Fig 19.
Fig. 19.
Neoascochyta cylindrispora (CBS 142456). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Latin cylindricus-, of cylindrical shape, and -spora, spore, due to the conidial morphology.
Description: Hyphae pale to dark brown, septate, smooth- and thin- to thick-walled, 4–6 μm wide. Conidiomata pycnidial, brown to dark brown, solitary or confluent, superficial on natural substrate (palm leaf), immersed in culture (OA), glabrous, subglobose, 150–300 × 130–160 μm, bearing a single ostiolar neck; pycnidial wall of textura angularis, composed of brown to dark brown, flattened polygonal cells of 4.5–11.5 μm diam, 2–4 layered, 15–60 μm thick,. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform or globose, 5 × 6 μm wide. Conidia 0–1-septate, hyaline, smooth- and thick-walled, mostly cylindrical or slightly allantoid, 11–11.5 × 3.5–4 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 30–34 mm diam after 7 d at 25 ± 1 °C, flattened, with an entire edge, dark green (M. 28F6); reverse dark green (M. 28F6) to greenish grey (M. 28F2). Colonies on MEA reaching 25–28 mm 7 d at 25 ± 1 °C, flattened, with an entire edge, white (M. 2A1) to olive grey (M. 2E2); reverse white (M. 2A1) to dark green (M. 27F3). NaOH spot test negative. Crystals absent. Optimal temperature for sporulation, 15 °C; optimal temperature of growth 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Materials examined: USA, from human corneal secretion, 2013, D.A. Sutton (holotype CBS H-23033, ex-holotype cultures CBS 142456 = UTHSC DI16-359 = FMR 13852); from human eye secretion, 2012, D.A. Sutton, culture UTHSC DI16-352 = FMR 13845.
Notes: Neoascochyta cylindrispora is phylogenetically distinct from N. desmazieri. It differs from the latter also morphologically by its glabrous pycnidia (covered by hyphal outgrowths in N. desmazieri), its smaller conidiogenous cells (5–6 μm wide vs. 7.5–11 μm wide in N. desmazieri) and shorter conidia (11–11.5 μm vs. 8.5–18 μm in N. desmazieri).
Neoascochyta desmazieri (Cavara) Q. Chen & L. Cai, Stud. Mycol. 82: 198. 2015.
Basionym: Ascochyta desmazieri Cavara, Z. Pflanzenkrankh 3: 21. 1893 (as “desmazieresii”).
Description: Chen et al. (2015).
Materials examined: Germany, Hohenlieth, from Lolium perenne, Apr. 1967, U.G. Schlösser (neotype HMAS 246690, ex-neotype living culture CBS 297.69). USA, from human respiratory tract, 2006, D.A. Sutton, living cultures UTHSC DI16-207 = FMR 13697; from unknow source of clinical sample, 2010, D.A. Sutton, living cultures UTHSC DI16-320 = FMR 13813; from human head superficial tissue sample, 2011, D.A. Sutton, living cultures UTHSC DI16-332 = FMR 13825; from human toe nail, 2011, D.A. Sutton, living cultures UTHSC DI16-341 = FMR 13834.
Notes: In this study four strains from human clinical specimens clustered with the ex-type strain of N. desmazieri, and those strains were morphologically and genetically identical with the type, only differing in location and substrate of isolation.
Neoascochyta tardicrescens Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, sp. nov., MycoBank MB819693. Fig 20.
Fig. 20.
Neoascochyta tardicrescens (CBS 689.97). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G, H. Conidiogenous cells. I. Conidia. Scale bars: F = 100 μm. G–I = 10 μm.
Etymology: From Latin tarde-, slowly, and -crescens, growing, in reference to the slow growing colonies.
Description: Hyphae pale to dark brown, septate, smooth- and thin- to thick-walled, 4–6 μm wide. Conidiomata pycnidial, brown to dark brown, solitary, superficial and immersed (OA), glabrous or covered with hyphal outgrows, globose to subglobose, 100–120 × 100–170 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–4 layered, composed of brown to dark brown, flattened polygonal cells of 12.5–25 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 5–10.5 × 5–8.5 μm. Conidia 1-septate, hyaline, smooth- and thick-walled, cylindrical to allantoid, 10–13.5 × 3–4 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 6 mm diam after 7 d at 25 ± 1 °C, flattened, undulate, dark green (M. 27F3); reverse olive brown (M. 4F3) to brownish grey (M. 4F2). Colonies on MEA reaching 7 mm diam after 7 d at 25 ± 1 °C, flattened, undulate, yellowhish grey (M. 2B2); reverse yellowish-brown (M. 5E8) to greenish grey (M. 28F2). NaOH spot test negative. Crystals absent. Optimal temperature for sporulation 15 °C; optimal temperature of growth 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Materials examined: Norway, Oslo, from hay, Apr. 1997, M. Torp (holotype CBS H-9005, ex-holotype living cultures CBS 689.97 = FMR 15352). USA, from human feet, 2010, D.A. Sutton, living cultures UTHSC DI16-291 = FMR 13783.
Notes: The strains CBS 689.97 and UTHSC DI16-291 grow and sporulate better at lower temperatures (around 15 °C) than at room temperature, and clearly differ morphologically from N. argentina in producing smaller conidiomata (100–120 × 100–170 μm vs. 210–390 × 140–270 ìm), in the presence of necks (absent vs. present), in the number of ostioles (1 vs. 1–3), and in their smaller conidiogenous cells (5–10.5 × 5–8.5 μm vs. 7.5–14.5 × 6–13.5 μm). Nonetheless, these strains formed a sister clade to N. argentina.
Clade C: Cucurbitariaceae G. Winter, Rabenhorst's Kryptogamen-Flora, Pilze-Ascomyceten 1.2: 308. 1885.
Type genus: Cucurbitaria Gray, Nat. Arr. Brit. Pl. (London) 1: 519. 1821.
Clade C1: Neocucurbitaria Wanas., E.B.G. Jones & K.D. Hyde, Mycosphere 8: 408. 2017.
Type species: Neocucurbitaria unguis-hominis (Punith. & M.P. English) Wanas. et al.
Neocucurbitaria aquatica Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, sp. nov. MycoBank MB822114.
Etymology: The species name refers to the habitat from which the fungus was recovered (sea water).
Culture sterile. Neocucurbitaria aquatica differs from its phylogenetically closest species N. unguis-hominis, based on the alignment of the concatenated four loci deposited in TreeBASE (S21115): LSU position, 412 (C); ITS positions, 539 (C), 595 (A); tub2 positions, 1121 (G), 1170 (G), 1257 (T); rpb2 positions, 1351 (A), 1387 (T), 1439 (T), 1801 (C), and 1816 (C).
Culture characteristics: Colonies on OA reaching 21–24 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 3F4); reverse olive (M. 3F4) to dark grey (M. 3F1). Colonies on MEA reaching 16–17 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish grey (M. 3B2); reverse grey (M. 3B1). NaOH spot test negative. Crystals absent. Optimal temperature of growth 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 35 °C.
Material examined: Montenegro, Kotor bay, from sea water, Oct. 1973, M. Muntañola-Cvetkovic, (holotype CBS H-16102, ex-holotype living culture CBS 297.74 = FMR 14867).
Notes: Neocucurbitaria aquatica was previously identified as “Pyrenochaeta quercina” based on LSU and SSU loci sequencing (de Gruyter et al. 2010). However, in our phylogenetic analysis using four loci, N. aquatica was closely related to Neocucurbitaria unguis-hominis. As N. aquatica was recovered from sea water, and is phylogenetically unrelated to the ex-type strain of Neocucurbitaria quercina (CBS 115095), we propose it as a new species.
Neocucurbitaria cava (Schulzer) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB821491. Fig. 21.
Fig. 21.
Neocucurbitaria cava (CBS 257.68). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiophores. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Phoma cava Schulzer, Verh. Zool.-bot. Ges. Wien 21:1248. 1871.
Synonyms: Aposphaeria cava (Schulzer) Sacc. & Schulzer, Syll. fung. (Abellini) 3: 174. 1884.
Coniothyrium cavum (Schulzer) Kuntze, Revis. gen. pl. (Leipzig) 3(2): 459. 1898.
Pleurophoma cava (Schulzer) Boerema et al., Persoonia 16: 172. 1996.
Pyrenochaeta cava (Schulzer) Gruyter et al., Mycologia 102: 1076. 2010.
Description from ex-epitype culture (CBS 257.68): Hyphae hyaline to brown, smooth- and thin-walled, septate, 2.5–3.5 μm wide. Conidiomata pycnidial, brown, solitary or confluent, semi-immersed or immersed (OA), glabrous, subglobose, 140–200 × 100–140 μm, with one ostiolar neck; pycnidial wall of textura angularis, composed of brown, flattened polygonal cells of 2.5–5 μm diam. Conidiophores hyaline, smooth-walled, straight or sinuous to slightly curved, slightly tapering towards the apex, branched at the base, 10–22 × 1.5–2.5 μm. Conidiogenous cells integrated to the conidiophore, phialidic, hyaline, smooth-walled, doliiform, with a more or less cylindrical collarette, up to 3 per conidiophore. Conidia aseptate, hyaline, smooth- and thin-walled, mostly cylindrical to slightly allantoid, 2.5–3.5 × 1–1.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 16 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 3F4); reverse dark grey (M. 3F1). Colonies on MEA reaching 14 mm after 7 d at 25 ± 1 °C, flattened, yellowish grey (M. 3B2); reverse olive brown (M. 4E4). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 35 °C.
Materials examined: Germany, Kiel-Kitzeberg, from wheat field soil, 1965, W. Gams (epitype CBS H-20320, ex-epitype living cultures CBS 257.68 = IMI 331911 = FMR 15747). Italy, on branch of Quercus cerris, M. Farras, living cultures CBS 115953 = FMR 15333.
Notes: Pyrenochaeta cava was epitypified by de Gruyter et al. (2010). In our phylogenetic analysis this species clustered in Neocucurbitaria, a genus recently introduced by Wanasinghe et al. (2017b). Therefore, we propose the new combination N. cava.
Neocucurbitaria hakeae (Crous) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB821492.
Basionym: Pyrenochaeta hakeae Crous, Persoonia 37: 353. 2016.
Description: Crous et al. (2016a).
Material examined: Australia, Western Australia, Denmark, Lights Beach, on leaves of Hakea sp., 19 Sep. 2015, P.W. Crous (holotype CBS H-22894, ex-holotype living cultures CBS 142109 = CPC 28920).
Notes: In our phylogenetic tree, this species forms a sister clade to N. cava. Therefore, we propose a new combination to accommodate this species in the genus Neocucurbitaria. Morphologically, N. hakeae resembles N. unguis-hominis, but the former species is the only species of the genus that produces pale brown conidiophores.
Neocucurbitaria irregularis Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819769. Fig. 22.
Fig. 22.
Neocucurbitaria irregularis (CBS 142791). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin irregularis, irregular, referring to the shape of its conidia.
Description: Hyphae brown, smooth- and thin-walled, septate, 2–5 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial (OA), glabrous, subglobose to ovoid, 75–130 × 65–120 μm, with 3–4 papillate ostiolar necks, pycnidial wall of textura angularis, 2–5 layered, 10–35 μm thick, composed of brown, flattened polygonal cells of 3–12 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, doliiform, 2.5 × 3.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, 2.5–4 × 1.5–2. μm, guttulate.
Culture characteristics: Colonies on OA reaching 17–18 mm diam after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4F6); reverse brownish grey (M. 4F2). Colonies on MEA reaching 11 mm after 7 d at 25 ± 1 °C, flattened, pale yellow (M. 4A3); reverse pale yellow (M. 4A4) to greyish yellow (M. 4C6). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 35 °C.
Material examined: USA, from human arm injury, 2000, D.A. Sutton (holotype CBS H-23029, ex-holotype living cultures CBS 142791 = UTHSC DI16-229 = FMR 13719).
Notes: Neocucurbitaria irregularis is proposed to accommodate a clinical isolate previously identified as “Pyrenochaeta unguis-hominis” (Valenzuela-Lopez et al. 2016). This isolate forms a basal clade together with N. keratinophila and N. unguis-hominis. However, it is morphologically well-differentiated from the latter two species, by having small, simple conidiogenous cells instead of filiform conidiophores.
Neocucurbitaria keratinophila (Verkley et al.) Valenzuela-Lopez, Stchigel, Guarro & Cano, comb. nov. MycoBank MB821494.
Basionym: Pyrenochaeta keratinophila Verkley et al., Revta Iberoamer. Micol. 27: 24. 2010.
Description: Verkley et al. (2010).
Material examined: Spain, Alicante, from human corneal scrapings (keratitis), Mar. 2007, A. Rodriguez & J. Guarro (holotype CBS H-20122, ex-holotype living CBS 121759 = FMR 9444).
Notes: This species described by Verkley et al. (2010) was isolated from a human corneal specimen with a case of keratitis. Morphologically it resembles Pyrenochaeta. However, in our phylogenetic analyses this species clustered close to N. irregularis. Therefore, we propose a new combination for this fungus in Neocucurbitaria.
Neocucurbitaria quercina (Kabát & Bubák) Wanas. et al., Mycosphere 8: 412. 2017. Fig. 23.
Fig. 23.
Neocucurbitaria quercina (CBS 115095). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Setae. H. Conidiophores. I. Conidia. Scale bars: F = 50 μm. G–I = 10 μm.
Basionym: Pyrenochaeta quercina Kabát & Bubák, Hedwigia 52: 342. 1912.
Description taken from Bubák & Kabát (1912), which is based on the holotype: Conidiomata pycnidial dark brown, solitary or confluent, setose, globose, 150–220 μm diam. Setae dark brown, tapered towards the apex, erect or decumbent, smooth- and thick-walled, up to 65 μm long, 5 μm broad at the base. Conidiophores cylindrical, tapered toward the apex, erect or slightly curved, hyaline, 25 × 3–3.5 μm. Conidia aseptate, hyaline, bacilliform, 2–3 × 1.5 μm.
Description from the ex-neotype culture (CBS 115095): Hyphae brown, smooth- and thin-walled, septate, 2.5–5 μm wide. Conidiomata pycnidial brown, solitary or confluent, superficial (OA), mostly glabrous or covered with somewhat shortest setae, globose, 70–90 μm diam, 100–230 × 90–130 μm when ovoid, with 1–2 papillate ostiolar necks; pycnidial wall of textura angularis, composed of brown, flattened polygonal cells of 3–12 μm diam; setae brown, erect, rounded at the top, septate, thin-walled, 7–10 × 2.5–3.5 μm,. Conidiophores hyaline, smooth-walled, straight or sinuous to slightly curved, slightly tapering towards the apex, branched at the base, 6.5–14 × 2–3 μm. Conidiogenous cells terminal and lateral on the conidiophore, phialidic, hyaline, smooth-walled, ampulliform when terminal, with a more or less cylindrical collarette, up to 4 per conidiophore. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to cylindrical, 1.5–3 × 1.2–1.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 21 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 3F4); reverse dark grey (M. 3F1). Colonies on MEA reaching 12 mm after 7 d at 25 ± 1 °C, flattened, olive (M. 3F4) to pale grey (M. 3B1); reverse dark grey (M. 3F1). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 35 °C.
Material examined: Italy, from Quercus robur, Nov 1971, S. Mutto Accordi (neotype designated here CBS H-23205, MBT377969, ex-neotype living cultures CBS 115095 = FMR 14868).
Notes: Bubák & Kabát (1912) described Pyrenochaeta quercina from Quercus cerris leaves, in Bukovina forest, Moldavia. The holotype is apparently missing. We studied the isolate CBS 115095, identified previously as P. quercina by de Gruyter et al. (2010), which has been recovered from Quercus robur in Italy. Recently, Wanasinghe et al. (2017b) transferred P. quercina to Neocucurbitaria. In our phylogenetic tree this strain clustered with N. cava and N. hakeae, confirming the right placement into Neocucurbitaria. Because the strain CBS 115095 was isolated from a related host to that of the basionym (both are different species of oaks), we designated this strain as the neotype for Pyrenochaeta quercina, in order to stabilize the taxonomy of the species.
Neocucurbitaria unguis-hominis (Punith. & M.P. English) Wanas. et al., Mycosphere 8: 412. 2017. Fig. 24.
Fig. 24.
Neocucurbitaria unguis-hominis (CBS 112.79). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G, H. Conidiophores. I. Conidia. Scale bars: F = 50 μm. G–I = 10 μm.
Basionym: Pyrenochaeta unguis-hominis Punith. & M.P. English, Trans. Br. mycol. Soc. 64: 539. 1975.
Description: Punithalingam & English (1975).
Materials examined: The Netherlands, Utrecht, from lung sample of Agapornis sp., C. Hoek, living cultures CBS 111112 = FMR 14866. USA, unknown substrate, 2006, D.A. Sutton, living cultures UTHSC DI16-213 = FMR 13703. Wales, Cardiff, from air sample, Apr. 1974, G.H. Boerema, living cultures CBS 112.79 = IMI 386095 = PD 74/1018 = FMR 15748.
Notes: Pyrenochaeta unguis-hominis was established by Punithalingam & English (1975) for a fungus recovered from a human toe-nail. Later, Wanasinghe et al. (2017b) considered this the type species of Neocucurbitaria. Interestingly, the three strains studied by us were able to grow and sporulate at 37 °C, being the only species of the genus that displays such abilities.
Clade C2: Paracucurbitaria
Paracucurbitaria Valenzuela-Lopez, Stchigel, Guarro & Cano, gen. nov. MycoBank MB821453.
Etymology: From Greek παρα-, beside, referring to the morphological similarity with the asexual morph of Cucurbitaria.
Conidiomata pycnidial, pale brown to brown, solitary or confluent, superficial or semi-immersed, pycnidial wall of textura angularis, 2–4 layered, glabrous or ornamented, subglobose to ovoid, ostiolate. Conidiophores if present, septate, hyaline, straight or sinuous to slightly curved, slightly tapering towards the apex. Conidiogenous cells integrated in the conidiophore, phialidic, hyaline, smooth-walled, ampulliform when terminal, with a more or less cylindrical collarette, several per conidiophore. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, guttulate.
Type species: Paracucurbitaria corni (Bat. & A.F. Vital) Valenzuela-Lopez, Stchigel, Guarro & Cano.
Paracucurbitaria corni (Bat. & A.F. Vital) Valenzuela-Lopez, Stchigel, Guarro & Cano, comb. nov. MycoBank MB821454. Fig. 25.
Fig. 25.
Paracucurbitaria corni (CBS 248.79). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium G. Conidiophores. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Plenodomus corni Bat. & A.F. Vital, Anais Soc. Biol. Pernambuco 15: 420. 1957.
Synonyms: Phoma riggenbachii Boerema & J.D. Janse, Eur. J. For. Path. 11: 428. 1981.
Pyrenochaeta corni (Bat. & A.F. Vital) Boerema, Loer. & Hamers, Persoonia 16: 158. 1996.
Description from reference strain (CBS 248.79): Hyphae hyaline to pale brown, smooth- and thin-walled, septate, 2.5–4 μm wide. Conidiomata pycnidial, pale brown to brown, solitary or confluent, superficial or semi-immersed (OA), glabrous, globose to subglobose, 110–210 × 110–190 μm diam, with 2–5 ostiolar necks; pycnidial wall of textura angularis, initially pseudoparenchymatous, scleroplectenchymatous with the age (mainly on MEA), 3–4 layered, 15–30 μm thick, composed of brown to dark brown, flattened polygonal cells of 3–6 μm diam. Conidiophores branched at the base, septate, hyaline, straight or sinuous to slightly curved, slightly tapering towards the apex, 6.5–18 μm long. Conidiogenous cells integrated in the conidiophore, phialidic, hyaline, smooth-walled, doliiform or ampulliform, 3.5–7.5 × 1.3–3.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, mostly cylindrical or rarely ovoid, 1.8–4 × 1.2–1.6 μm, guttulate.
Culture characteristics: Colonies on OA reaching 14 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 2E6); reverse olive (M. 2E6) to dark grey (M. 2F1). Colonies on MEA reaching 10 mm after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4D6) to dark grey (M. 4F1); reverse olive brown (M. 4D6) to dark grey (M. 4F1). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined The Netherlands, Scheerwolde, from Fraxinus excelsior with bacterial canker (also from Prays fraxinella), 1978, deposited by G. H. Boerema, living cultures CBS 248.79 = PD 78/1092 = FMR 16593.
Notes: Plenodomus corni was erected by Batista & Vital (1957) as a new species on branches of Cornus sanguinea from Hungary, and it was characterised by producing brown to black, solitary or clustered, mostly immersed, glabrous, globose to subglobose, pycnidial conidiomata of 115–135 μm diam, with a pseudoparenchymatous wall 12.5–20 μm thick, composed of polygonal to subglobose cells of 2.5–4 μm diam, with phialidic, hyaline, filiform or flask-shaped conidiogenous cells, 3.5–6 × 1–2 μm, and hyaline, bacilliform, 1.5–3 × 1 μm conidia. Later, Janse (1981) isolated a similar fungus from Fraxinus excelsior with a bacterial canker, and also from dead, discoloured tissue surrounding galleries and holes of Prays fraxinella (ash bud moth). This fungus (living culture CBS 248.79) was considered by Boerema et al. (1981) as the same taxon as Plenodomus corni. However, a new name was necessary to transfer Plenodomus corni to the genus Phoma because the species name was occupied (Phoma corni Fuckel ex Sacc.). The strain CBS 248.79 was characterised by the production of pycnidial conidiomata with a scleroplectenchymatous wall, variable in size and in shape, 100–200 μm diam, and of aseptate conidia (measuring 2.1–2.6 × 0.8–1.2 μm), produced on elongated conidiogenous cells. However, CBS 248.79 shows some morphological variation depending of the culture media employed: on MEA it shows a scleroplectenchymatous wall as given in the original description by Janse, but on OA it resembles the description given by Batista & Vital (1957), but it does not produce setose pycnidia as mentioned by Boerema et al. (1996). The strain CBS 248.79 forms a distinct monophyletic clade within the Cucurbitariaceae. Therefore, we propose the new combination, Paracucurbitaria corni.
Paracucurbitaria italica Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, sp. nov. MycoBank MB822116. Fig. 26.
Fig. 26.
Paracucurbitaria italica (CBS 234.92). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiophores. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: The name of the species refers to the country of origin of the fungus, Italy.
Description: Hyphae hyaline to pale brown, smooth- and thin-walled, septate, 2.5–4 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial or semi-immersed (OA), covered by hyphal outgrowths, subglobose to ovoid, 190–240 × 170–190 μm diam, with 1–2 ostiolar necks; pycnidial wall of textura angularis, 2–4 layered, 10–15 μm thick, composed of brown to dark brown, flattened polygonal cells of 5–13 μm diam. Conidiophores septate, hyaline, straight or sinuous to slightly curved, slightly tapering towards the apex, 15–20 μm long. Conidiogenous cells phialidic, hyaline, smooth-walled, filiform or flask-shaped, 4–9 × 2–3.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ellipsoidal to cylindrical, 2.5–3 × 1–1.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 13 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 2E6); reverse olive (M. 2E6) to dark grey (M. 2F1). Colonies on MEA reaching 11 mm after 7 d at 25 ± 1 °C, flattened, white (M. 2A1); reverse white (M. 2A1). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: Italy, Rende, from Olea europaea leaves, 26 Feb. 1992, C. Candiano (holotype CBS H-16104, ex-holotype living cultures CBS 234.92 = FMR 14869).
Notes: The strain CBS 234.92 was previously identified as “Pyrenochaeta corni” by de Gruyter et al. (2010). However, this strain is phylogenetically distinct from its closest relative, Paracucurbitaria corni, and differs morphologically by the production of ornamented conidiomata (covered with hyphal outgrowths vs. glabrous). Consequently, we propose CBS 234.92 as the ex-type strain of Paracucurbitaria italica sp. nov.
Clade C3: Allocucurbitaria
Allocucurbitaria Valenzuela-Lopez, Stchigel, Guarro & Cano, gen. nov. MycoBank MB821455.
Etymology: From Greek àλλο-, different, due to is related but phylogenetically and morphologically different to the genus Cucurbitaria.
Conidiomata pycnidial, brown, solitary or confluent, superficial, pycnidial wall of textura angularis, glabrous, subglobose to ovoid, ostiolate. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical to allantoid, guttulate.
Type species: Allocucurbitaria botulispora Valenzuela-Lopez, Stchigel, Guarro & Cano.
Allocucurbitaria botulispora Valenzuela-Lopez, Stchigel, Guarro & Cano, sp. nov. MycoBank MB819770. Fig. 27.
Fig. 27.
Allocucurbitaria botulispora (CBS 142452). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiophores. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin botulus-, sausage, and -spora, spores, due to the shape of the conidia.
Description: Hyphae pale brown, smooth- and thin-walled, septate, 1.5–2.5 μm wide. Conidiomata pycnidial, brown, confluent, superficial (OA), glabrous, subglobose to ovoid, 60–160 × 60–120 μm diam, with 1–2 papillate ostiolar necks; pycnidial wall of textura angularis, 2–4 layered, 10–30 μm thick, composed of pale brown to brown, flattened polygonal cells of 3–10 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 5–8 × 2–2.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical to allantoid, 3–5 × 1–1.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 26–29 mm diam after 7 d at 25 ± 1 °C, flattened, greyish yellow (M. 4C6); reverse olive brown (M. 4D5). Colonies on MEA reaching 22 mm after 7 d at 25 ± 1 °C, slightly floccose, yellowish white (M. 4A2); reverse pale orange (M. 5A3) to deep orange (M. 5A8). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 37 °C.
Material examined: USA, from human scab on leg, 2009, D.A. Sutton (holotype CBS H-23028, ex-holotype living cultures CBS 142452 = UTHSC DI16-273 = FMR 13764).
Notes: The strain CBS 142452 (= UTHSC DI16-273) was originally assigned to Pyrenochaeta (Valenzuela-Lopez et al. 2016). Morphologically, this strain displays a morphology more similar to phoma-like taxa (with glabrous pycnidia) than to species of Pyrenochaeta (because of its setose conidiomata). In our phylogenetic analysis, this fungus was placed in an uncertain taxonomic position within Cucurbitariaceae. Therefore, we proposed to accommodate CBS 142452 as a new species of the new genus Allocucurbitaria.
Clade D: Pseudopyrenochaetaceae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, fam. nov. MycoBank MB820426.
Etymology: From Latin pseudo-, resembling but not equalling, because the morphological similarity to Pyrenochaeta.
Conidiomata pycnidial, brown to dark brown, solitary, setose, globose to subglobose, papillate, ostiolate. Conidiophores simple, filiform, septate. Conidiogenous cells phialidic, intercalary, disposed along the conidiophores as short side projections. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical to allantoid.
Type genus: Pseudopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Pseudopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, gen. nov. MycoBank MB820427.
Etymology: The name refers to the morphological similarity with the genus Pyrenochaeta.
Conidiomata pycnidial, brown to dark brown, solitary, setose, globose to subglobose, with a papillate ostiolar neck. Conidiophores hyaline, simple, filiform, septate. Conidiogenous cells phialidic, hyaline, intercalary along the conidiophore, arising as very short lateral projections immediately below the transverse septa. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical to allantoid.
Type species: Pseudopyrenochaeta lycopersici (R.W. Schneid. & Gerlach) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Pseudopyrenochaeta lycopersici (R.W. Schneid. & Gerlach) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820431.
Basionym: Pyrenochaeta lycopersici R.W. Schneid. & Gerlach, Phytopath. Z. 56: 121. 1966.
Description: Schneider & Gerlach (1966).
Material examined: Germany, Berlin, from Lycopersicon esculentum root, Nov. 1971, R. Schneider & G.H. Boerema (isotype CBS H-17628, ex-isotype culture CBS 306.65 = FMR 15746 = BBA 9911 = DSM 62931).
Notes: In previous studies, the ex-isotype strain of Pyrenochaeta lycopersici (CBS 306.65) was phylogenetically located in the Cucurbitariaceae (de Gruyter et al. 2010, Wanasinghe et al. 2017b). However, de Gruyter et al. (2013) placed it as incertae sedis. According to our results, P. lycopersici falls phylogenetically outside the family Cucurbitariaceae and represents a new genus, Pseudopyrenochaeta, in the new family, Pseudopyrenochaetaceae.
Pseudopyrenochaeta terrestris Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, sp. nov. MycoBank MB822117.
Etymology: The species name refers to soil, the substrate from which the fungus was recovered.
Culture sterile. Pseudopyrenochaeta terrestris differs from its closest phylogenetic species, P. lycopersici, based on the alignment of the concatenated four loci deposited in TreeBASE (S21115), by six bp of LSU, 20 bp of ITS, 16 bp of tub2, and 47 bp of rpb2.
Culture characteristics: Colonies on OA reaching 22 mm diam after 7 d at 25 ± 1 °C, flattened, olive grey (M. 3E3); reverse olive grey (M. 3E3) to dark grey (M. 3F1). Colonies on MEA reaching 11 mm after 7 d at 25 ± 1 °C, slightly flattened, white (M. 3A1); reverse yellowish grey (M. 3C2). NaOH spot test negative. Crystals absent.
Material examined: The Netherlands, Naaldwijk, from greenhouse soil, Feb. 1972, L.H. Kaastra-Höweler (holotype FMR H-15327, ex-holotype living cultures CBS 282.72 = FMR 15327).
Notes: The strain CBS 282.72, deposited as “Pyrenochaeta lycopersici”, clustered with the ex-type strain of Pseudopyrenochaeta lycopersici. However, both strains differ significantly in all nucleotide sequences of the phylogenetic markers used in the present study. Therefore, strain CBS 282.72 is proposed here as the new species Pseudopyrenochaeta terrestris.
Clade E: Neopyrenochaetaceae Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, fam. nov. MycoBank MB820416.
Etymology: Relating to the distinct phenotypic and genetic relationship to the genus Pyrenochaeta and its relatives.
Conidiomata pycnidial, pale brown to brown, solitary, pycnidial wall of textura angularis, setose, ovoid to globose, with a non-papillate or papillate ostiolar neck. Conidiogenous cells phialidic, ampulliform or lageniform. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to subcylindrical.
Type genus: Neopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Neopyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, gen. nov. MycoBank MB820313.
Etymology: Referring to its morphological similarity to the genus Pyrenochaeta.
Conidiomata pycnidial, pale brown to brown, solitary, pycnidial wall of textura angularis, setose, ovoid to globose, ostiolate. Conidiogenous cells phialidic, ampulliform or lageniform. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to subcylindrical.
Type species: Neopyrenochaeta acicola (Moug. & Lév.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Neopyrenochaeta acicola (Moug. & Lév.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820314. Fig. 28.
Fig. 28.
Neopyrenochaeta acicola (CBS 812.95). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G, H. Conidiogenous cells. I. Conidia. Scale bars: F = 50 μm. G–I = 10 μm.
Basionym: Vermicularia acicola Moug. & Lév. apud Léveillé, Annls Sci. nat. (Bot.) III, 9:259. 1848 (as “Moug. Lév.”; non Phoma acicola sensu Saccardo), Syll. Fung. 3:100. 1884 [as “(Lév.) Sacc.”; = Sclerophoma pythiophila (Corda) Höhn.].
Synonym: Phoma leveillei var. leveillei Boerema & G.J. Bollen, Persoonia 8: 115. 1975.
Description and synonymy: Boerema et al. (2004).
Material examined: The Netherlands, from water pipe sample, 1995, Y. Driessen (neotype CBS H-20314, ex-neotype living cultures CBS 812.95 = FMR 14872).
Notes: Pyrenochaeta acicola was neotypified and relegated to the Cucurbitariaceae by de Gruyter et al. (2010). Although Neopyrenochaeta acicola morphologically resembles a Pyrenochaeta species, our phylogenetic analyses revealed that this taxon is distant from the type species of Pyrenochaeta, P. nobilis, and therefore we proposed the new genus Neopyrenochaeta for this and a few related species.
Neopyrenochaeta fragariae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, sp. nov. MycoBank MB820316. Fig. 29.
Fig. 29.
Neopyrenochaeta fragariae (CBS 101634). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: Relating to the host from the fungus was isolated, Fragaria (strawberry).
Description: Hyphae pale brown, smooth- and thin-walled, septate, 2.5–3 μm wide. Conidiomata pycnidial, pale brown to brown, solitary, superficial (OA), ovoid to globose, 170–220 × 160–210 μm, covered with brown to dark brown, septate, erect, smooth- and thick-walled setae tapering towards the apex, 110–120 × 2.5–5.5 μm, mainly disposed around the ostiole, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–5 layered, 20–60 μm thick, composed of brown, flattened polygonal cells of 5–15 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 4.5–7 × 3.5–4 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to ellipsoidal, 3.5–5 × 2–3 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 14 mm diam after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4E8); reverse olive brown (M. 4F2). Colonies on MEA reaching 11 mm after 7 d at 25 ± 1 °C, flattened, yellowish-brown (M. 5F4); reverse yellowish brown (M. 5E4). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: The Netherlands, Arnhem, from Fragaria sp., 1976, M.M.J. Dorenbosch (holotype CBS H-23206, ex-holotype living cultures CBS 101634 = PD 76/416 = FMR 14871).
Notes: The strain CBS 101634 was previously named Pyrenochaeta acicola. Although it is morphologically similar to the latter mentioned species (now in Neopyrenochaeta), these fungi differ in 23 and 11 nucleotides for rpb2 and tub2, respectively. Therefore, a new species name is proposed for CBS 101634.
Neopyrenochaeta inflorescentiae (Crous et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820317.
Basionym: Pyrenochaeta inflorescentiae Crous et al., CBS Diversity Ser. (Utrecht) 7: 115. 2008.
Description: Marincowitz et al. (2008).
Material examined: South Africa, Western Cape Province, from Protea neriifolia, 6 Jun. 2000, S. Marincowitz (holotype PREM 58657, ex-holotype living cultures CBS 119222 = CPC 13163 = FMR 15334).
Notes: In our phylogenetic analysis, the ex-type strain of Pyrenochaeta inflorescentiae (CBS 119222) clustered with N. acicola and N. fragariae in a terminal clade distant from the type species of the genus Pyrenochaeta, P. nobilis, and outside the family Cucurbitariaceae, where that fungus was previously placed. For that reason, we accommodate P. inflorescentiae in the new genus Neopyrenochaeta (Neopyrenochaetaceae).
Neopyrenochaeta telephoni (Rohit Sharma et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820318.
Basionym: Pyrenochaeta telephoni Rohit Sharma et al., Persoonia 35: 321. 2015.
Description: Crous et al. (2015b).
Material examined: India, Maharashtra, Pune, from screen of mobile phone, 2013, R. Kurli & P. Rahi (holotype MCC H1001, ex-holotype living cultures MCC 1159 = CBS 119222 = FMR 15754).
Notes: Recently, Sharma et al. (in Crous et al. 2015b) proposed the new species Pyrenochaeta telephoni, recovered from a mobile phone. Morphologically, it resembles other species of Pyrenochaeta; however, in our phylogenetic analysis this fungus forms a basal terminal clade in Neopyrenochaeta, which is distant from Cucurbitariaceae s. str.
Clade F: Pyrenochaetopsidaceae Valenzuela-Lopez, Crous, Cano, Guarro & Stchigel, fam. nov. MycoBank MB820308.
Conidiomata pycnidial, pale brown to brown, solitary or confluent; pycnidial wall of textura angularis, glabrous or setose, subglobose to ovoid, with a non-papillate or papillate ostiolar neck. Conidiogenous cells phialidic, hyaline, discrete or integrated in septate, acropleurogenous conidiophores. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid, cylindrical to allantoid, guttulate.
Type genus: Pyrenochaetopsis Gruyter, Aveskamp & Verkley.
Clade F1: Pyrenochaetopsis
Pyrenochaetopsis Gruyter, Aveskamp & Verkley, Mycologia 102: 1076. 2010.
Conidiomata pycnidial, honey to citrine or olivaceous to olivaceous black, solitary to confluent, superficial or submerged, with a non-papillate or papillate ostiolar neck; pycnidial wall pseudoparenchymatous, setose, globose to subglobose. Conidiogenous cells phialidic, hyaline, discrete and integrated in septate, acropleurogenous conidiophores. Conidia aseptate, cylindrical to allantoid, guttulate (de Gruyter et al. 2010).
Type species: Pyrenochaetopsis leptospora (Sacc. & Briard) Gruyter et al.
Pyrenochaetopsis americana Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB822115.
Etymology: The species name denotes the geographic area where the fungus is from.
Culture sterile. Pyrenochaetopsis americana differs from its closest phylogenetic species, Pyrenochaetopsis uberiformis, in five nucleotides for ITS, 19 for tub2 and 34 for rpb2, based on alignment of the concatenated four loci deposited in TreeBASE (S21115).
Culture characteristics: Colonies on OA reaching 30 mm diam after 7 d at 25 ± 1 °C, flattened, dark olive (M. 3F3); reverse olive grey (M. 3D2). Colonies on MEA reaching 19 mm diam after 7 d at 25 ± 1 °C, flattened, olive grey (M. 3D2) to white (M. 3A1); reverse white (M. 3A1). NaOH spot test negative. Crystals absent.
Material examined: USA, substrate unknown, 2007, D.A. Sutton (holotype FMR H-13715, ex-holotype living cultures UTHSC DI16-225 = FMR 13715).
Notes: The strain UTHSC DI16-225, which remained sterile in all culture media tested in this study, forms an unsupported sister clade with P. uberiformis, from which it is phylogenetically distant. Therefore, UTHSC DI16-225 is proposed here as a new species different from P. uberiformis.
Pyrenochaetopsis botulispora Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819764. Fig. 30.
Fig. 30.
Pyrenochaetopsis botulispora (CBS 142458). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Latin botulus-, sausage, and -spora, spore, relating to the morphology of the conidia.
Description: Hyphae brown, smooth- and thin-walled, septate, 2–7 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial (OA), glabrous, subglobose or globose, 140–190 × 130–160 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–3 layered, 15–35 μm thick, composed of brown, flattened polygonal cells of 5–8 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, subglobose, ca. 4 × 5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 4.5–6 × 2–2.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 25–30 mm diam after 7 d at 25 ± 1 °C, flattened, with abundant production of pycnidia, yellowish brown (M. 5E8); reverse yellowish-brown (M. 5F6). Colonies on MEA reaching 30 mm diam after 7 d at 25 ± 1 °C, flattened, orange grey (M. 5B2) to brownish orange (M. 5C5); reverse yellowish brown (M. 5E7) to greyish orange (M. 5B5). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human sputum sample, 2011, D.A. Sutton (holotype CBS H-23035, ex-holotype living cultures CBS 142458 = UTHSC DI16-298 = FMR 13791); from human bronchial wash sample, 2010, D.A. Sutton, living culture UTHSC DI16-289 = FMR 13781; from human foot skin, 2011, D.A. Sutton, living culture UTHSC DI16-297 = FMR 13790.
Notes: Pyrenochaetopsis botulispora is proposed to accommodate three isolates from clinical specimens, which form a sister clade to P. paucisetosa, being well differentiated phylogenetically from their closest relatives. Morphologically, P. botulispora is characterised by producing glabrous pycnidia, which are setose in P. paucisetosa, and by its slightly longer conidia (4.5–6 × 2–2.5 μm vs. 3–4 × 2–2.5 μm in in P. paucisetosa).
Pyrenochaetopsis confluens Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819763. Fig. 31.
Fig. 31.
Pyrenochaetopsis confluens (CBS 142459). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin confluens, confluent, due to the production of tightly aggregated conidiomata.
Description: Hyphae pale brown, smooth- and thin-walled, septate, 2–5 μm wide. Conidiomata pycnidial, pale brown, translucent, aggregated, immersed (MEA), subglobose or globose, 80–140 × 70–90 μm, with 1–2 papillate ostiolar necks, covered by brown setae around the ostiole; setae erect, smooth- and thick- walled, septate, 15–22.5(–35) × 2.5–4.5 μm; pycnidial wall of textura angularis, 2–3 layered, 13–20 μm thick, composed of brown, flattened polygonal cells of 5–8 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, subglobose, 4.5–7.5 × 6.5–7.5 μm. Conidia aseptate, hyaline, aseptate, smooth- and thin-walled, guttulate, ovoid to cylindrical, 2–4 × 2–2.5 μm.
Culture characteristics: Colonies on OA reaching 15 mm diam after 7 d at 25 ± 1 °C, flattened, white (M. 4A1) to olive brown (M. 4E4); reverse olive brown (M. 4F3). Colonies on MEA reaching 10 mm diam after 7 d at 25 ± 1 °C, flattened, white (M. 4A1) to brownish-grey (M. 4F2); reverse brownish-grey (M. 4F2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human blood sample, 2011, D.A. Sutton (holotype CBS H-23036, ex-holotype living cultures CBS 142459 = UTHSC DI16-303 = FMR = 13796).
Notes: The strain CBS 142459 forms a distinct clade phylogenetically distant from P. decipiens and P. indica. This new species grows slowly on all culture media tested and produces aggregated conidiomata.
Pyrenochaetopsis decipiens (Marchal) Gruyter et al., Mycologia 102: 1077. 2010.
Basionym: Pyrenochaeta decipiens Marchal, Bull. Soc. Roy. Bot. Belg. 30:139. 1891.
Synonym: Phoma terricola Boerema, Versl. Meded. plziektenk. Dienst Wageningen 163 (Jaarb. 1984): 38. 1985.
Material examined: The Netherlands, Hoofddorp, on cyst of Globodera pallida, May 1985, D. Hugo, No. 727 (neotype CBS H-20315, ex-neotype living cultures CBS 343.85 = IMI 386097 = FMR 14880).
Notes: In this study, newer genomic sequences data from the ex-type strain of Pyrenochaetopsis decipiens are provided. Unfortunately, we have not been able to induce this fungus to sporulate.
Pyrenochaetopsis globosa Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB821496. Fig. 32.
Fig. 32.
Pyrenochaetopsis globosa (CBS 143034). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin globosus, globose, due to the production of globose conidiomata.
Description: Hyphae hyaline to pale brown, smooth- and thin-walled, septate, 2–4 μm wide. Conidiomata pycnidial, pale olivaceus-brown to brown, solitary or aggregated, semi-immersed or immersed, mainly globose (70–200 μm diam), sometimes ovoid (150–220 × 140–190 μm), glabrous or covered by hyphal outgrowths, with 1–2 papillate ostiolar necks; pycnidial wall of textura angularis, 3–5 layered, 25–35 μm thick, composed of pale olive-brown to brown, flattened polygonal cells of 3–10 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, lageniform to ampulliform, 3.5–5 × 2.5–3 μm. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to cylindrical, 3–5.5 × 1.5–2 μm, guttulate.
Culture characteristics: Colonies on OA reaching 27 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish brown (M. 5E5); reverse greyish brown (M. 5F3). Colonies on MEA reaching 20 mm diam after 7 d at 25 ± 1 °C, flattened, brownish-orange (M. 5C3); reverse pale brown (M. 5D4). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human dermatitis sample, 2009, D.A. Sutton (holotype CBS H-23208 ex-holotype living cultures CBS 143034 = UTHSC DI16-275 = FMR 13766).
Notes: The strain CBS 143034, which is morphologically similar to P. uberiformis (slightly different in pycnidial and conidial size), forms a large clade wherein there is P. uberiformis and several other species of the genus Pyrenochaetopsis. Because the nucleotide sequences of both fungi differ in 19 bp for rpb2 and 13 bp for tub2, P. globosa is proposed as a new species for the genus.
Pyrenochaetopsis indica (T.S. Viswan.) Gruyter et al., Mycologia 102: 1077. 2010.
Basionym: Pyrenochaeta indica T.S. Viswan., Curr. Sci. 26:118. 1957.
Synonym: Phoma indica (T.S. Viswan.) Gruyter & Boerema, Persoonia 17: 556. 2002.
Description: Boerema et al. (2004).
Material examined: India, Poona, on leaf spot of Saccharum officinarum (holotype AMH-11, ex-holotype living cultures IMI 062569 = CBS 124454 = FMR 14879).
Notes: We studied the ex-type strain of Pyrenochaetopsis indica, providing new genomic sequence data. It is morphologically characterised by its setose pycnidia, and the production of globose to subglobose, olivaceous chlamydospores solitary or in chains. Morphologically it is difficult to differentiate this species from P. decipiens. However, Pyrenochaetopsis indica clearly differs genetically from the latter in its tub2 and rpb2 sequences. Unfortunately, all cultures remained sterile.
Pyrenochaetopsis leptospora (Sacc. & Briard) Gruyter et al., Mycologia 102: 1076. 2010. Fig. 33.
Fig. 33.
Pyrenochaetopsis leptospora (CBS 101635). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Pyrenochaeta leptospora Sacc. & Briard, Revue Mycol. 11: 16. 1889.
Synonyms: Pyrenochaeta spegazziniana Trotter, Syll. Fung. 25: 190. 1931.
Phoma briardii Gruyter & Boerema, Persoonia 17: 555. 2002.
Description: Boerema et al. (2004).
Material examined: Germany, subtrate unknown, J.W. Veenbaas, living cultures CBS 122787 = FMR 14873. The Netherlands, on Secale cereal (epitype CBS H-20313, ex-epitype living cultures CBS 101635 = PD 71/1027 = FMR 14877).
Notes: We received isolate CBS 122787 as “Coniothyrium cerealis”, but it was identified as P. leptospora in our phylogenetic study.
Pyrenochaetopsis microspora (Gruyter & Boerema) Gruyter et al., Mycologia 102: 1077. 2010. Fig. 34.
Fig. 34.
Pyrenochaetopsis microspora (CBS 102876). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Phoma leveillei var. microspora Gruyter & Boerema, Persoonia 17: 553. 2002.
Description: Boerema et al. (2004).
Materials examined: Montenegro, Lake of Skadar, from water, 1975 (holotype HLB 999-242399, ex-holotype living cultures CBS 102876 = PD 75/911 = FMR 14874). USA, from human sinusitis sample, 2006, D.A. Sutton, living cultures UTHSC DI16-193 = FMR 13688.
Notes: In this paper, the ex-type strain of Pyrenochaetopsis microspora was examined, and new genomic sequence data and illustrations are provided. Furthermore, one human clinical specimen clustered with the ex-type living culture, being morphologically and genetically very closely related.
Pyrenochaetopsis paucisetosa Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819766. Fig. 35.
Fig. 35.
Pyrenochaetopsis paucisetosa (CBS 142460). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin paucus, few, and -setosus, setose, because the conidiomata are covered by a few setae.
Description: Hyphae brown, smooth- and thin-walled, septate, 2–3 μm wide. Conidiomata pycnidial, brown, solitary, superficial or immersed (OA), setose, globose to ovoid, 150–190 × 140–160 μm, with a papillate ostiolar neck, covered by a few, brown, erect or slightly curved, smooth- and thick-walled, septate setae, (50–)63–68(–83) × 2–3.5 μm; pycnidial wall of textura angularis, 2–5 layered, 20–50 μm thick, composed of brown, flattened polygonal cells of 4–13 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 3.5–4 × 3–3.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 3–4 × 2–2.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 25 mm diam after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4F5); reverse brownish grey (M. 4F2). Colonies on MEA reaching 21 mm diam after 7 d at 25 ± 1 °C, floccose, pale grey (M. 4C1); reverse medium-grey (M. 4E1). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 35 °C.
Material examined: USA, from human toe nail, 2005, D.A. Sutton (holotype CBS H-23037, ex-holotype living cultures CBS 142460 = UTHSC DI16-193 = FMR 13683).
Notes: Pyrenochaetopsis paucisetosa, recovered from a specimen of superficial human tissue, produces pycnidia covered by a few setae, and conidia smaller than in other species of the genus. Phylogenetically, P. paucisetosa is well-separated from P. botulispora.
Pyrenochaetopsis poae Crous & Quaedvlieg, Persoonia 32: 197. 2014.
Description: Crous et al. (2014).
Material examined: Netherlands, Raalte, on Poa sp. (Poaceae), 2013, W. Quaedvlieg (holotype CBS H-21677, ex-holotype living cultures CBS 136769 = D779 = FMR 14876).
Notes: We studied the ex-type strain of Pyrenochaetopsis poae, which is morphologically similar to the generic type of P. leptospora. In this paper, we provide rpb2 sequence that, together with tub2, are useful to differentiate these taxa.
Pyrenochaetopsis setosissima Valenzuela-Lopez, Cano, Crous, Guarro & Stchigel, sp. nov. MycoBank MB819767. Fig. 36.
Fig. 36.
Pyrenochaetopsis setosissima (CBS 119739). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Etymology: From Latin –setosissimus, bearing many setae, relating to the ornamentation of the pycnidia.
Description: Hyphae brown, smooth- and thin-walled, septate, 2–5 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial (OA), subglobose to ovoid, 150–230 × 150–200 μm, with a papillate ostiolar neck, covered by many dark brown, erect, smooth- and thick-walled, septate setae, 33–83 × 2–4 μm; pycnidial wall of textura angularis, 2–4 layered, 20–50 μm thick, composed of brown, flattened polygonal cells of 5–15 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 5–4.5 × 4–4.5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 4–5 × 2–2.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 25 mm diam after 7 d at 25 ± 1 °C, flattened, olive brown (M. 4F4); reverse brownish grey (M. 4F2). Colonies on MEA reaching 18 mm diam after 7 d at 25 ± 1 °C, flattened, light orange (M. 5A4); reverse orange white (M. 5A2). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: Brazil, Minas Gerais, Lavras, from Coffea arabica leaf, Jun. 1999, L.H. Pfenning (holotype CBS H-23209, ex-holotype living cultures CBS 119739 = FMR 14875).
Notes: The isolate CBS 119739 was identified as P. microspora by de Gruyter et al. (2010) using SSU and LSU sequences as phylogenetic markers. However, in our phylogenetic study employing more markers, it clusters distant from the latter species. Pyrenochaetopsis setosissima is morphologically very similar to P. microspora, and can only be distinguished based on molecular data (differing in 19 bp for tub2 and 31 bp for rpb2).
Pyrenochaetopsis uberiformis Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB819765. Fig. 37.
Fig. 37.
Pyrenochaetopsis uberiformis (CBS 142461). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: From Latin –ubera, mammaries, and -forma, shape, relating to the anatomy of its pycnidia.
Description: Hyphae brown, smooth- and thin-walled, septate, 2–3 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial or immersed (OA), glabrous, globose or ovoid, 200–440 × 130–410 μm, with a papillate ostiolar neck; pycnidial wall of textura angularis, 2–4 layered, 15–30 μm thick, composed of pale brown to brown, flattened polygonal cells of 5–10 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 3–4 × 4–5 μm. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, 4–6 × 2–2.5 μm, guttulate.
Culture characteristics: Colonies on OA reaching 27 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish brown (M. 5E5); reverse greyish brown (M. 5F3). Colonies on MEA reaching 20 mm diam after 7 d at 25 ± 1 °C, flattened, brownish orange (M. 5C3); reverse pale brown (M. 5D4). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human ear lesion, 2009, D.A. Sutton (holotype CBS H-23038, ex-holotype living cultures CBS 142461 = UTHSC DI16-277 = FMR 13769).
Notes: The strain CBS 142461 clustered within the Pyrenochaetopsis clade, distant from other species of the genus, with the exception of P. americana, which forms a sister clade. Both strains differ in their rpb2 and tub2 sequences. Therefore, we propose strain CBS 142461 as representative of the new species P. uberiformis.
Clade F2: Xenopyrenochaetopsis
Xenopyrenochaetopsis Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, gen. nov. MycoBank MB820311.
Etymology: From Greek ξένος-, strange, alien, because it is phylogenetically distinct from the genus Pyrenochaetopsis.
Conidiomata pycnidial, pale brown to brown, solitary or confluent; pycnidial wall of textura angularis, glabrous, globose, ostiolate. Conidiogenous cells phialidic, hyaline. Conidia aseptate, hyaline, smooth- and thin-walled, cylindrical, guttulate.
Type species: Xenopyrenochaetopsis pratorum (P.R. Johnst. & Boerema) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Xenopyrenochaetopsis pratorum (P.R. Johnst. & Boerema) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820312. Fig. 38.
Fig. 38.
Xenopyrenochaetopsis pratorum (CBS 445.81). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium. G. Conidiogenous cells. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Basionym: Phoma pratorum P.R. Johnst. & Boerema, New Zealand J. Bot. 19: 395. 1981.
Synonym: Pyrenochaetopsis pratorum (P.R. Johnst. & Boerema) Gruyter et al., Stud. Mycol. 75: 24. 2012.
Description from ex-isotype (CBS 445.81): Hyphae pale brown to brown, smooth- and thin-walled, septate, 2.5–5 μm wide. Conidiomata pycnidial, pale brown to brown, solitary or confluent, semi-immersed or immersed (OA), glabrous, globose to irregular, (88–)160–270 × (80–)100–250 μm, with 1–3 papillate ostiolar necks; pycnidial wall of textura angularis, 2–4 layered, 10–30 μm thick, composed of pale brown to brown, flattened polygonal cells of 2.5–8 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 3–3.5 × 1.5–2 μm. Conidia aseptate, hyaline, smooth- and thin-walled, subreniform to oblong or cylindrical, 4–5 × 1.5–2 μm, guttulate.
Culture characteristics: Colonies on OA reaching 5 mm diam after 7 d at 25 ± 1 °C, flattened, olive (M. 3D4) to olive grey (M.3F2); reverse olive (M.3F4). Colonies on MEA reaching 4 mm diam after 7 d at 25 ± 1 °C, flattened, yellowish white (M. 3A2); reverse ash blonde (M. 3C3). NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 15 °C; maximum temperature of growth 25 °C.
Material examined: New Zealand, Rakura, near Hamilton, from a leaf of Lolium perenne (Poaceae), 1980, P.R. Johnston (isotype CBS H-7625, CBS H-7626, ex-isotype living cultures CBS 445.81 = PDDCC 7049 = PD 80/1254 = FMR 14878).
Notes: Pyrenochaetopsis pratorum was proposed as a new combination for Phoma pratorum by de Gruyter et al. (2013). In that study, it clustered with Pyrenochaetopsis but was situated phylogenetically distinct from P. leptospora. However, in our phylogenetic analysis this species clustered outside Pyrenochaetopsis s. str. Moreover, Phoma pratorum differs in the main distinctive morphological feature of the genus Pyrenochaetopsis, the production of setose pycnidia (glabrous in P. pratorum). Therefore, we accommodate this species in the new genus Xenopyrenochaetopsis.
Clade F3: Neopyrenochaetopsis
Neopyrenochaetopsis Valenzuela-Lopez, Cano, Guarro & Stchigel, gen. nov. MycoBank MB820309.
Etymology: Referring to its close phylogenetic relationship with the genus Pyrenochaetopsis.
Conidiomata pycnidial, brown, solitary or confluent, pycnidial wall of textura angularis, glabrous, subglobose to ovoid, ostiolate. Conidiogenous cells phialidic, ampulliform to globose. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to cylindrical.
Type species: Neopyrenochaetopsis hominis Valenzuela-Lopez, Cano, Guarro & Stchigel.
Neopyrenochaetopsis hominis Valenzuela-Lopez, Cano, Guarro & Stchigel, sp. nov. MycoBank MB820310. Fig. 39.
Fig. 39.
Neopyrenochaetopsis hominis (CBS 143033). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cell. H. Conidia. Scale bars: F = 100 μm. G, H = 10 μm.
Etymology: Relating to its isolation from a human specimen.
Description: Hyphae pale yellow to pale brown, smooth- and thin-walled, septate, 2–3 μm wide. Conidiomata pycnidial, brown, solitary or confluent, superficial or immersed (OA), glabrous, subglobose to ovoid, 160–170 × 140–160 μm, with a single papillate ostiolar neck; pycnidial wall of textura angularis, 2–4 layered, 15–40 μm thick, composed of brown, flattened polygonal cells of 2.5–8 μm diam. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform to globose, 4–5 μm diam wide. Conidia aseptate, hyaline, smooth- and thin-walled, ovoid to narrowly ellipsoidal, 3–3.5 × 1.5–2 μm, guttulate. Chlamydospores absent.
Culture characteristics: Colonies on OA reaching 31 mm diam after 7 d at 25 ± 1 °C, flattened, greyish yellow (M. 3B4); reverse greyish yellow (M. 3C4); yellow pigment diffusing into the agar. Colonies on MEA reaching 29 mm diam after 7 d at 25 ± 1 °C, floccose, dull yellow (M. 3B3) to white (M. 3A1); reverse brownish yellow (M. 5C8); diffusible pigment yellowish. NaOH spot test negative. Crystals absent. Optimal temperature of growth and sporulation 25 °C; minimum temperature of growth 5 °C; maximum temperature of growth 30 °C.
Material examined: USA, from human skin tissue, 2007, D. A. Sutton (holotype CBS H-23207, culture ex-holotype living cultures CBS 143033 = UTHSC DI16-238 = FMR 13728).
Notes: The strain CBS 143033, recovered from a clinical sample, forms a distinct basal clade within the Pyrenochaetopsidaceae. Morphologically, N. hominis can be differentiated from the other taxa mainly by the production of smaller-sized conidia, and a yellow diffusing pigment on MEA and OA.
Clade N: Parapyrenochaetaceae Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, fam. nov. MycoBank MB820418.
Etymology: Named after its close morphological relationship with Pyrenochaeta.
Conidiomata pycnidial, brown, solitary, pycnidial wall of textura angularis, setose, globose, ostiolate. Conidiogenous cells phialidic, ampulliform or lageniform. Conidia aseptate, hyaline, smooth- and thin-walled, allantoid or ellipsoidal.
Type genus: Parapyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Parapyrenochaeta Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, gen. nov. MycoBank MB820319.
Etymology: Based on its close morphological relationship to Pyrenochaeta.
Conidiomata pycnidial, pale brown to brown, solitary, setose, globose, ostiolate; pycnidial wall of textura angularis. Conidiogenous cells phialidic, ampulliform or lageniform. Conidia aseptate, hyaline, smooth- and thin-walled, allantoid or ellipsoidal.
Type species: Parapyrenochaeta protearum (Crous) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano.
Parapyrenochaeta acaciae (Crous et al.) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820321. Fig. 40.
Fig. 40.
Parapyrenochaeta acaciae (CBS 141291). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidia. G. Conidiogenous cells. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Pyrenochaeta acaciae Crous et al., Persoonia 36: 349. 2016.
Description: Crous et al. (2016b).
Material examined: Australia, Victoria, on leaf of Acacia sp. (Fabaceae), 7 Nov. 2014, J. Edwards, I.G. Pascoe & P.W. Crous (holotype CBS H-22601, ex-holotype living cultures CPC 25527 = CBS 141291 = FMR 15755).
Notes: Pyrenochaeta acaciae was described by Crous et al. (2016b) based on morphological and nucleotide sequence data, highlighting the close relationship with P. protearum. In our phylogenetic study, P. acaciae clustered distant from the Cucurbitariaceae s. str., forming a distinct clade related to P. protearum.
Parapyrenochaeta protearum (Crous) Valenzuela-Lopez, Crous, Stchigel, Guarro & Cano, comb. nov. MycoBank MB820320. Fig. 41.
Fig. 41.
Parapyrenochaeta protearum (CBS 131315). A, B. Colony on OA (front and reverse). C, D. Colony on MEA (front and reverse). E. Pycnidia forming on OA. F. Pycnidium G. Conidiophores. H. Conidia. Scale bars: F = 50 μm. G, H = 10 μm.
Basionym: Pyrenochaeta protearum Crous, Persoonia 27: 153. 2011.
Synonym: Pyrenochaeta pinicola Crous, Persoonia 32: 255. 2014.
Description: Crous et al. (2011).
Materials examined: France, Nice, L’aire d’Esterel petrol filling station, on needles of Pinus sp., 20 Jul. 2013, P.W. Crous, living cultures ex-type of P. pinicola, CPC 23455 = CBS 137997 = FMR 15753. South Africa, Western Cape Province, on leaves of Protea mundii, 4 May 2010, P.W. Crous (holotype of P. protearum, CBS H-20772, ex-holotype living cultures CPC 18322 = CBS 131315 = FMR 15752).
Notes: Pyrenochaeta protearum morphologically resembles phoma-like taxa in producing single phialides covering the inner source of the pycnidia, and having small, ((3–)4–5(–6) × (2–)2.5(–3) μm), aseptate, hyaline conidia, but also resembles pyrenochaeta-like species due to its setose pycnidia (Crous et al. 2011). Based on ITS and LSU nucleotide sequences, this fungus has been related to Leptosphaeria, Pyrenochaeta and Pyrenochaetopsis, and was included in the genus Pyrenochaeta (Crous et al. 2011). However, our results revealed that this fungus is phylogenetically distant from Pyrenochaeta spp., and from the members of the Cucurbitariaceae, and therefore we accommodated it in the new genus Parapyrenochaeta. We also studied the ex-type strain of Pyrenochaeta pinicola (Crous et al. 2014), which was morphologically and genetically very closely related to Pa. protearum. Therefore, we reduce Py. pinicola to synonymy under Pa. protearum.
Discussion
The taxonomy of the coelomycetes has undergone major changes in recent years, mainly due to the extensive use of molecular techniques, which has resulted in a more natural classification of these fungi. In this regard, the taxonomic circumscription of the genera Phoma (Didymellaceae) and Pyrenochaeta (Cucurbitariaceae) have proven to be especially complex. In recent studies on Didymellaceae, Chen et al., 2015, Chen et al., 2017 restricted Phoma to P. herbarum, accepting 17 genera in the family Didymellaceae. They demonstrated that by combining four loci, but especially by using the rpb2 marker, it was possible to resolve the phylogeny of the Didymellaceae. However, in recent studies, several Phoma species accepted by Aveskamp et al. (2010) such as P. bulgarica, P. crystallifera, P. destructiva, P. eupyrena, P. multirostrata, P. omnivirens, P. pereupyrena and P. saxea, were not included. Currently, several genera such as Didymellocamarosporium, Endocoryneum, Heracleicola, Neodidymella, Platychora and Pseudohendersonia have been added to the Didymellaceae based mainly on the ribosomal gene analyses (Ariyawansa et al., 2015a, Hyde et al., 2016, Wijayawardene et al., 2016). However, recently, Chen et al. (2017) demonstrated that the genera mentioned above are simple synonyms of previous genera of that family such as Ascochyta, Boeremia, Stagonosporopsis and Neomicrosphaeropsis. Therefore, sequences of those taxa need to be verified with proper genes to resolve their taxonomic placement within the Didymellaceae. For this reason, our proposal was to revise this family testing a large set of coelomycetous fungi recently isolated from clinical specimens (Valenzuela-Lopez et al. 2016), but also including several reference species of Phoma from the prior study of Aveskamp et al. (2010). This resulted in the proposal of six new genera, viz. Cumuliphoma, Ectophoma, Juxtiphoma, Remotididymella, Similiphoma and Vacuiphoma, 14 new species, and nine new combinations.
The taxonomic placement of Pyrenochaeta continues to be a topic of discussion, as this genus accommodates at least 163 epithets (www.indexfungorum.org). It is currently related to the Cucurbitariaceae, but an earlier phylogenetic study involving Pyrenochaeta species performed by Schoch et al. (2006), showed that P. nobilis, its type species, occupied an unclear taxonomic placement within the Pleosporales. Subsequently, this genus occupied an intermediate position as incertae sedis between the Leptosphaeriaceae and Didymellaceae (de Gruyter et al. 2009), or belonging to the Leptosphaeriaceae (Zhang et al. 2009). Later, de Gruyter et al. (2010) placed Pyrenochaeta in Cucurbitariaceae, and several species of Phoma in the new genus Pyrenochaetopsis. However, by employing additional gene loci in our phylogeny, the type species P. nobilis clustered distant from Cucurbitariaceae s. str., being placed as incertae sedis in the Pleosporineae. Moreover, several species previously identified as Pyrenochaeta have proved to be phylogenetically scattered within the Pleosporineae. Therefore, we introduced four new families with several new genera to accommodate all Pyrenochaeta species which clustered outside the Cucurbitariaceae, i.e. Neopyrenochaetaceae (which includes Neopyrenochaeta gen. nov.), Parapyrenochaetaceae (within Parapyrenochaeta gen. nov.), Pseudopyrenochaetaceae (including Pseudopyrenochaeta gen. nov.) and Pyrenochaetopsidaceae (including the two new genera, Neopyrenochaetopsis and Xenopyrenochaetopsis).
In the revision of Cucurbitariaceae by Doilom et al. (2013), the authors accepted six genera in the family, although Curreya, Rhytidiella and Syncarpella were not sequenced. This family was recently enlarged by Wanasinghe et al. (2017b) proposing the new genus Neocucurbitaria to accommodate N. acerina, N. unguis-hominis (syn. Pyrenochaeta unguis-hominis, the type species of that genus) and N. quercina (syn. Pyrenochaeta quercina) and considering the genus Fenestella as belonging to this family; however, the type species and more species of this genus should be studied to clarify its taxonomy. Neocucurbitaria has been also modified in our study to include N. cava (syn. Pyrenochaeta cava), N. hakeae (syn. Pyrenochaeta hakeae), N. keratinophila (syn. Pyrenochaeta keratinophila), and the new species N. aquatica and N. irregularis. Here, we have also enlarged the current concept of Cucurbitariaceae with the proposal of the new genera Allocucurbitaria (with the only species A. botulispora), which is closely related to Cucurbitaria and Paracucurbitaria, with P. corni and the new species P. italica forming a clade distinct from Neocucurbitaria. In fact, Cucurbitariaceae is currently circumscribed with four genera, i.e. the three mentioned above, and Cucurbitaria. In contrast, Camarosporium, which was included in Cucurbitariaceae by Doilom et al. (2013), has been recently placed in Coniothyriaceae by Crous & Groenewald (2017), who studied and epitypified the generic type of Camarosporium and several phoma-like species, proposing the new family Libertasomycetaceae within Pleosporineae. In the same year, Wanasinghe et al. (2017a) have studied a large set of camarosporium-like fungi proposing the new families Camarosporidiellaceae and Neocamarosporiaceae and resurrected the family Camarosporiaceae. However, in our phylogeny, several members of Coniothyriaceae and Leptosphaeriaceae remain in an ambiguous taxonomic position within Pleosporineae. Furthermore, in our study the family Camarosporidiellaceae was phylogenetically unsupported, which is probably caused by the lack of rpb2 or tub2 sequences; therefore further studies are needed to understand the relationships of this family with the other members of this suborder.
At the present study, we have clarified the generic concept of two of the largest genera of coelomycetes (Phoma and Pyrenochaeta) through a polyphasic approach that included the analysis of four phylogenetic markers of 143 additional isolates. This approach allowed a better delimitation of members of Cucurbitariaceae and Didymellaceae of the suborder Pleosporineae that currently encompasses the following 19 families: Camarosporiaceae, Camarosporidiellaceae, Coniothyriaceae, Cucurbitariaceae, Didymellaceae, Dothidotthiaceae, Halojulellaceae, Leptosphaeriaceae, Libertasomycetaceae, Microsphaeropsidaceae, Neocamarosporiaceae, Neophaeosphaeriaceae, Neopyrenochaetaceae, Parapyrenochaetaceae, Phaeosphaeriaceae, Pleosporaceae, Pseudopyrenochaetaceae, Pyrenochaetopsidaceae and Shiraiaceae.
Acknowledgements
The authors are very grateful to the following researchers or institutions: Markéta Šandová (Curator of the Natural History Museum, Czech Republic, PRM), Fátima Sales (Curator of the Herbarium of the Botanical Garden of the University of Coimbra, Portugal), Marc Jeanson and Simon Chagnoux (Muséum National d'Histoire Naturelle, Paris, France), Begoña Aguirre-Hudson (Curator of the Royal Botanic Gardens, Kew, UK), Rossella Marcucci (Curator of the Saccardo’s Herbarium, Università degli Studi di Padova, Italy) and Jamie Minnaert-Grote and Andrew Miller (The Illinois Natural History Survey Fungarium, Illinois, USA). We also thank to reviewers for their valuable comments and suggestions to improve the content of the manuscript. This study was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL2013-43789-P.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
References
- Ahmed S.A., Desbois N., Quist D. Phaeohyphomycosis caused by a novel species, Pseudochaetosphaeronema martinelli. Journal of Clinical Microbiology. 2015;53:2927–2934. doi: 10.1128/JCM.01456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.A., Hofmüller W., Seibold M. Tintelnotia, a new genus in Phaeosphaeriaceae harbouring agents of cornea and nail infections in humans. Mycoses. 2017;60:244–253. doi: 10.1111/myc.12588. [DOI] [PubMed] [Google Scholar]
- Ahmed S.A., Van De Sande W.W., Stevens D.A. Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia. 2014;33:141–154. doi: 10.3767/003158514X684744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaradasa B.S., Madrid H., Groenewald J.Z. Porocercospora seminalis gen. et comb. nov., the causal organism of buffalograss false smut. Mycologia. 2014;106:77–85. doi: 10.3852/13-147. [DOI] [PubMed] [Google Scholar]
- Ariyawansa H.A., Hyde K.D., Jayasiri S.C. Fungal diversity notes 111–252–taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2015;75:27–274. [Google Scholar]
- Ariyawansa H.A., Jones E.B.G., Suetrong S. Halojulellaceae a new family of the order Pleosporales. Phytotaxa. 2013;130:14–24. [Google Scholar]
- Ariyawansa H.A., Phukhamsakda C., Thambugala K.M. Revision and phylogeny of Leptosphaeriaceae. Fungal Diversity. 2015;74:19–51. [Google Scholar]
- Aveskamp M.M., de Gruyter J., Crous P.W. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Diversity. 2008;31:1–18. [Google Scholar]
- Aveskamp M.M., de Gruyter J., Woudenberg J.H.C. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveskamp M.M., Verkley G.J.M., de Gruyter J. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia. 2009;101:363–382. doi: 10.3852/08-199. [DOI] [PubMed] [Google Scholar]
- Batista A.C., Vital A.F. Contribuiçao ao Estudo dos Fungos Sphaeropsidales. Anais da Sociedade de Biologia de Pernambuco. 1957;15:413–427. [Google Scholar]
- Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – II. Section Peyronellaea. Persoonia. 1993;15:197–221. [Google Scholar]
- Boerema G.H., Bollen G.J. Conidiogenesis and conidial septation as differentiating criteria between Phoma and Ascochyta. Persoonia. 1975;8:111–444. [Google Scholar]
- Boerema G.H., de Gruyter J., Noordeloos M.E. Contributions towards a monograph of Phoma (Coelomycetes) – IV. Section Heterospora: taxa with large sized conidial dimorphs, in vivo sometimes as Stagonosporopsis synanamorphs. Persoonia. 1997;16:335–371. [Google Scholar]
- Boerema G.H., de Gruyer J., Noordeloos M.E. CABI Publishing; Wallingford, UK: 2004. Phoma identification manual. Differentiation of specific and infra-specific taxa in culture. [Google Scholar]
- Boerema G.H., Dorenbosch M.M.J., Kesteren H.A. van. Remarks on species of Phoma referred to Peryonellaea. Persoonia. 1965;4:47–68. [Google Scholar]
- Boerema G.H., Kesteren H.A. van, Loerakker W.M. Notes on Phoma. Transactions of the British Mycological Society. 1981;77:61–74. [Google Scholar]
- Boerema G.H., Loerakker W.M., Hamers M.E. Contributions towards a monograph of Phoma (Coelomycetes) – III. 2. Misapplications of the type species name and the generic synonyms of section Plenodomus (Excluded species) Persoonia. 1996;16:141–189. [Google Scholar]
- Borman A.M., Desnos-Ollivier M., Campbell C.K. Novel taxa associated with human fungal black-grain mycetomas: Emarellia grisea gen. nov., sp. nov., and Emarellia paragrisea sp. nov. Journal of Clinical Microbiology. 2016;54:1738–1745. doi: 10.1128/JCM.00477-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunaud P. Supplément à la liste des Sphaeroidées trouvées à Saintes. Revue Mycologique Toulouse. 1887;9:13–17. [Google Scholar]
- Bubák F., Kabát J.E. Mykologische Beiträge. VII. Hedwigia. 1912;52:340–363. [Google Scholar]
- Chandra S., Tandon R.N. Three new leaf-infecting fungi from Allahabad. Mycopathologia et Mycologia Applicata. 1966;29:273–276. [Google Scholar]
- Chen Q., Hou L.W., Duan W.J. Didymellaceae revisited. Studies in Mycology. 2017;87:105–159. doi: 10.1016/j.simyco.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Jiang J.R., Zhang G.Z. Resolving the Phoma enigma. Studies in Mycology. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda A.C.J. Vol. 4. 1840. pp. 1–53. (Icones fungorum hucusque cognitorum). [Google Scholar]
- Crous P.W., Carris L.M., Giraldo A. The Genera of Fungi – fixing the application of the type species of generic names – G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus. 2015;6:163–198. doi: 10.5598/imafungus.2015.06.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Groenewald J.Z. They seldom occur alone. Fungal Biology. 2016;120:1392–1415. doi: 10.1016/j.funbio.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Crous P.W., Groenewald J.Z. The Genera of Fungi – G 4: Camarosporium and Dothiora. IMA Fungus. 2017;8:131–152. doi: 10.5598/imafungus.2017.08.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Shivas R.G., Quaedvlieg W. Fungal Planet description sheets: 214–280. Persoonia. 2014;32:184–306. doi: 10.3767/003158514X682395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Summerell B.A., Shivas R.G. Fungal Planet description sheets: 92–106. Persoonia. 2011;27:130–162. doi: 10.3767/003158511X617561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2009. Fungal Biodiversity. CBS Laboratory Manual Series. [Google Scholar]
- Crous P.W., Wingfield M.J., Burgess T.I. Fungal Planet description sheets: 469–557. Persoonia. 2016;37:218–403. doi: 10.3767/003158516X694499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Le Roux J.J. Fungal Planet description sheets: 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P.W., Wingfield M.J., Richardson D.M. Fungal Planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.L., Currah R.S. Atradidymella muscivora gen. et sp. nov. (Pleosporales) and its anamorph Phoma muscivora sp. nov.: a new pleomorphic pathogen of boreal bryophytes. American Journal of Botany. 2009;96:1281–1288. doi: 10.3732/ajb.0900010. [DOI] [PubMed] [Google Scholar]
- De Gruyter J. Contributions towards a monograph of Phoma (Coelomycetes) – IX Section Macrospora. Persoonia. 2002;18:85–102. [Google Scholar]
- De Gruyter J., Aveskamp M.M., Woudenberg J.H.C. Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycological Research. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- De Gruyter J., Boerema G.H. Contributions towards a monograph of Phoma (Coelomycetes) – VIII. Section Paraphoma: taxa with setose pycnidia. Persoonia. 2002;17:541–561. [Google Scholar]
- De Gruyter J., Boerema G.H., van der Aa H.A. Contributions towards a monograph of Phoma (Coelomycetes) VI – 2. Section Phyllostictoides: outline of its taxa. Persoonia. 2002;18:1–53. [Google Scholar]
- De Gruyter J., Noordeloos M.E. Contributions towards a monograph of Phoma (Coelomycetes) – I. 1. Section Phoma: taxa with very small conidia in vitro. Persoonia. 1992;15:71–92. [Google Scholar]
- De Gruyter J., Woudenberg J.H., Aveskamp M.M. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- De Gruyter J., Woudenberg J.H.C., Aveskamp M.M. Redisposition of Phoma-like anamorphs in Pleosporales. Studies in Mycology. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog G.S., Guarro J., Gené J., Figueras M.J. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2011. Atlas of clinical fungi. CD-ROM v. 3.1. [Google Scholar]
- Doilom M., Liu J.K., Jaklitsch W.M. An outline of the family Cucurbitariaceae. Sydowia. 2013;65:167–192. [Google Scholar]
- Dorenbosch M.M.J. Key to nine ubiquitous soil-borne Phoma-like fungi. Persoonia. 1970;6:1–14. [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher N.L., Burgess L.W., Toussoun T.A., Nelson P.E. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology. 1982;72:151–153. [Google Scholar]
- Hashimoto A., Matsumura M., Hirayama K. Pseudodidymellaceae fam. nov.: phylogenetic affiliations of mycopappus-like genera in Dothideomycetes. Studies in Mycology. 2017;87:187–206. doi: 10.1016/j.simyco.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M., Gené J., Castañeda-Ruiz R.F. Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology. 2017;86:53–97. doi: 10.1016/j.simyco.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne A.S. Diagnoses of fungi from “spotted” apples. Journal of Botany, British and Foreign. 1920;58:238–242. [Google Scholar]
- Hyde K.D., Hongsanan S., Jeewon R. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity. 2016;80:1–270. [Google Scholar]
- Hyde K.D., Jones E.B.G., Liu J.K. Families of Dothideomycetes. Fungal Diversity. 2013;63:1–313. [Google Scholar]
- Jaklitsch W.M., Checa J., Blanco M.N. A preliminary account of the Cucurbitariaceae. Studies in Mycology. 2017 doi: 10.1016/j.simyco.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W.M., Olariaga I., Voglmayr H. Teichospora and the Teichosporaceae. Mycological Progress. 2016;15:31. doi: 10.1007/s11557-016-1171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W.M., Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Studies in Mycology. 2016;85:35–64. doi: 10.1016/j.simyco.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W.M., Voglmayr H. Three former taxa of Cucurbitaria and considerations on Petrakia in the Melanommataceae. Sydowia. 2017;69:81–95. doi: 10.12905/0380.sydowia69-2017-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse J.D. The bacterial disease of ash (Fraxinus excelsior), caused by Pseudomonas syringae subsp. savastanoi pv. Fraxini II. Etiology and taxonomic considerations. European Journal of Forest Pathology. 1981;11:425–438. [Google Scholar]
- Kirk P.M., Cannon P.F., Minter D.W., Stalpers J.A. 10th ed. CAB International; Wallingford, UK: 2008. Ainsworth and Bisby's Dictionary of the Fungi. [Google Scholar]
- Kocakaya Z., Halici M.G., Kocakaya M. Phoma candelariellae sp. nov., a lichenicolous fungus from Turkey. Mycotaxon. 2015;130:1185–1189. [Google Scholar]
- Kornerup A., Wanscher J.H. 3rd ed. Methuen; London, England: 1978. Methuen Handbook of Colour. [Google Scholar]
- Kruys A., Eriksson O.E., Wedin M. Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycological Research. 2006;110:527–536. doi: 10.1016/j.mycres.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Lawrey J.D., Diederich P., Nelsen M.P. Phylogenetic placement of lichenicolous Phoma species in the Phaeosphaeriaceae (Pleosporales, Dothideomycetes) Fungal Diversity. 2012;55:195–213. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Liu Y.X., Hyde K.D., Ariyawansa H.A. Shiraiaceae, new family of Pleosporales (Dothideomycetes, Ascomycota) Phytotaxa. 2013;103:51–60. [Google Scholar]
- Marincowitz S., Crous P.W., Groenewald J.Z., Wingfield M.J. Westerdijk Fungal Biodiversity Institute; Utrecht, The Netherlands: 2008. Microfungi Occurring on Proteaceae in the Fynbos. [CBS Biodiversity Series No. 7] [Google Scholar]
- Matsushima T. Matsushima mycological memoirs 7. Matsushima Mycological Memoirs. 1993;7:1–141. [Google Scholar]
- McDonald G.K., Peck D. Effects of crop rotation, residue retention and sowing time on the incidence and survival of ascochyta blight and its effect on grain yield of field peas (Pisum sativum L.) Field Crops Research. 2009;111:11–21. [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond: 1–8. Association for Computing Machinery; USA: 2012. The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. [Google Scholar]
- Noordeloos M.E., de Gruyter J., van Eijk G.W., Roeijmans H.J. Production of dendritic crystals in pure cultures of Phoma and Ascochyta and its value as a taxonomic character relative to morphology, pathology and cultural characteristics. Mycological Research. 1993;97:1343–1350. [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University; Sweden: 2004. MrModeltest v2. Program Distributed by the Author. [Google Scholar]
- Plowright C.B. On the fungoid diseases of the tomato. Gardeners' Chronicle. 1881;16:620–622. [Google Scholar]
- Punithalingam E. Sphaeropsidales in culture from humans. Nova Hedwigia. 1979;31:119–158. [Google Scholar]
- Punithalingam E. 1985. Phoma sorghina. CMI Descriptions of Pathogenic Fungi and Bacteria No. 825. [Google Scholar]
- Punithalingam E., English M.P. Pyrenochaeta unguis-hominis sp. nov. on human toe-nails. Transactions of the British Mycological Society. 1975;64:539–541. [Google Scholar]
- Rai M.K. Phoma sorghina infection in human being. Mycopathologia. 1989;105:167–170. doi: 10.1007/BF00437250. [DOI] [PubMed] [Google Scholar]
- Rehner S.A., Samuels G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research. 1994;98:625–634. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel T., Balesdent M.H. The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Molecular Plant Pathology. 2005;6:225–241. doi: 10.1111/j.1364-3703.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- Saccardo P.A. Fungi Gallici lecti a cl. viris P. Brunaud, C.C. Gillet et Abb. Letendre. Michelia. 1879;1:500–538. [Google Scholar]
- Saccardo P.A. Vol. 10. Supplementum Universale, Pars II; Italy, Padova: 1892. pp. 1–964. (Sylloge Fungorum omnium hucusque cognitorum). [Google Scholar]
- Salam M.U., MacLeod W.J., Maling T. A meta-analysis of severity and yield loss from ascochyta blight on field pea in Western Australia. Australasian Plant Pathology. 2011;40:591–600. [Google Scholar]
- Schneider R., Gerlach W. Pyrenochaeta lycopersici nov. spec., der Erreger der Korkwurzelkrankheit der Tomate. Journal of Phytopathology. 1966;56:117–122. [Google Scholar]
- Schoch C.L., Shoemaker R.A., Seifert K.A. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia. 2006;98:1041–1052. doi: 10.3852/mycologia.98.6.1041. [DOI] [PubMed] [Google Scholar]
- Sharma R., Sharma R., Crous P.W. Matsushimamyces, a new genus of keratinophilic fungi from soil in central India. IMA Fungus. 2015;6:337–343. doi: 10.5598/imafungus.2015.06.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spegazzini C. Nova addenda ad mycologiam venetam. Atti della Società Crittogamologica Italiana. 1881;3:42–71. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.Y., Qi Y.L., Cai L. Induction of sporulation in plant pathogenic fungi. Mycology. 2012;3:195–200. [Google Scholar]
- Sung G.-H., Sung J.-M., Hywel-Jones N.L. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolutionary. 2013;31:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Hirayama K., Yonezawa H. Revision of the Massarineae (Pleosporales, Dothideomycetes) Studies in Mycology. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambugala K.M., Daranagama D.A., Phillips A.J.L. Microfungi on Tamarix. Fungal Diversity. 2016;82:239–306. [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakunyingcharoen T., Lombard L., Groenewald J.Z. Mycoparasitic species of Sphaerellopsis, and allied lichenicolous and other genera. IMA Fungus. 2014;5:391–414. doi: 10.5598/imafungus.2014.05.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Lopez N., Sutton D.A., Cano-Lira J.F. Coelomycetous fungi in the clinical setting: morphological convergence and cryptic diversity. Journal of Clinical Microbiology. 2016;55:552–567. doi: 10.1128/JCM.02221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkley G.J., Gené J., Guarro J. Pyrenochaeta keratinophila sp. nov., isolated from an ocular infection in Spain. Revista Iberoamericana de Micología. 2010;27:22–24. doi: 10.1016/j.riam.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe D.N., Hyde K.D., Crous P.W. Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Studies in Mycology. 2017;87:207–256. doi: 10.1016/j.simyco.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe D.N., Jones E.B., Camporesi E. Taxonomy and phylogeny of Laburnicola gen. nov. and Paramassariosphaeria gen. nov. (Didymosphaeriaceae, Massarineae, Pleosporales) Fungal Biology. 2016;120:1354–1373. doi: 10.1016/j.funbio.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Wanasinghe D.N., Phookamsak R., Jeewon R. Fenestellaceae with descriptions of new Fenestella species and Neocucurbitaria gen. nov. Mycosphere. 2017;8:397–414. [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego, California, USA: 1990. pp. 315–322. [Google Scholar]
- Wijayawardene N.N., Crous P.W., Kirk P.M. Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Diversity. 2014;69:1–55. doi: 10.1007/s13225-014-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene N.N., Hyde K.D., Wanasinghe D.N. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Diversity. 2016;77:1–316. [Google Scholar]
- Winter G. Vol. 1. 1885. Pilze–Ascomyceten; pp. 65–528. (GL Rabenhorst's Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz). [Google Scholar]
- Woudenberg J.H.C., Aveskamp M.M., de Gruyter J. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg J.H.C., Groenewald J.Z., Binder M. Alternaria redefined. Studies in Mycology. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Crous P.W., Schoch C.L., Hyde K.D. Pleosporales. Fungal Diversity. 2012;53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Schoch C.L., Fournier J. Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re–evaluation. Studies in Mycology. 2009;64:85–102. doi: 10.3114/sim.2009.64.04. [DOI] [PMC free article] [PubMed] [Google Scholar]