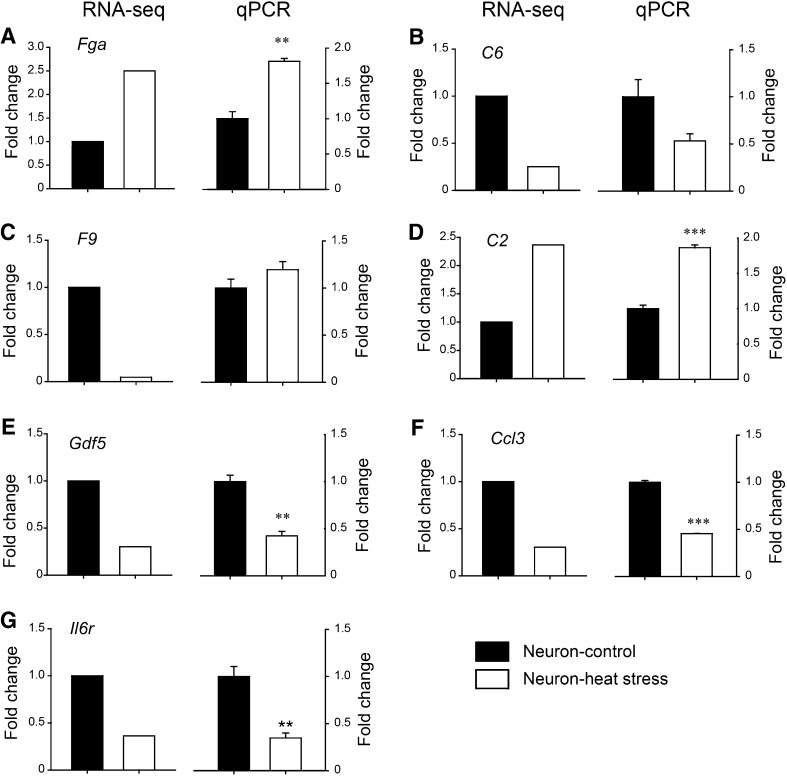
Fig. 5.
Validation of DEGs in heat-stressed neurons. Selected DEGs in neurons were detected by qPCR (n = 3/group) to validate the results of RNA-seq. Left panels: fold changes of RNA-seq; right panels: fold changes in qPCR.: A **P = 0.0017 for Fga, B P = 0.076 for C6, C P = 0.200 for F9, D ***P < 0.001 for C2, E **P = 0.0022 for Gdf5, F ***P < 0.001 for Ccl3, and G **P = 0.0054 for Il6r (Student’s t-test).