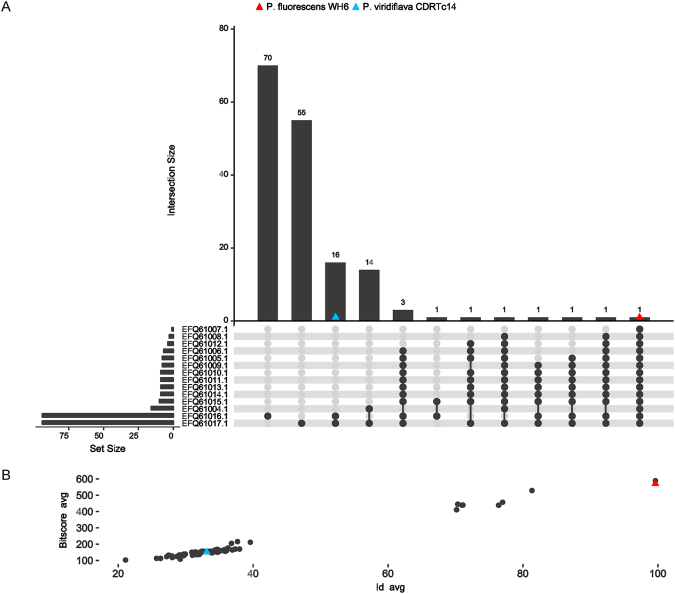
Figure 4.
Occurrence of formylaminooxyvinylglycine (FVG) biosynthetic genes (gvg gene cluster of WH6) across tested genomes are shown by UpSet composite plot: (A) Intersection size vs set size: this plot shows the number of genomes (vertical barplots) sharing the same combination of gvg cluster elements (black dots read vertically) whereas the horizontal barplots report how many genomes have one specific element of the gvg cluster, gene by gene (black dots read horizontally). As expected, all the genes of the gvg cluster are present in P. fluorescens WH6 genome (red triangle). None of the other Pseudomonas genomes has the complete set of elements of the cluster (at least with an identity percentage ≥ 75%). P. viridiflava CDRTc14 shows only two genes and these are also the most abundant among Pseudomonas genomes. (B) Scatterplot showing the identity % average (x-axis) and the bitscore average (y-axis) for all the genes in the gvg cluster in each genome. P. fluorescens WH6 has an id avg of 100% and bitscore avg of 590, P. viridiflava CDRTc14 has an id avg of 32% and bitscore avg of 136.