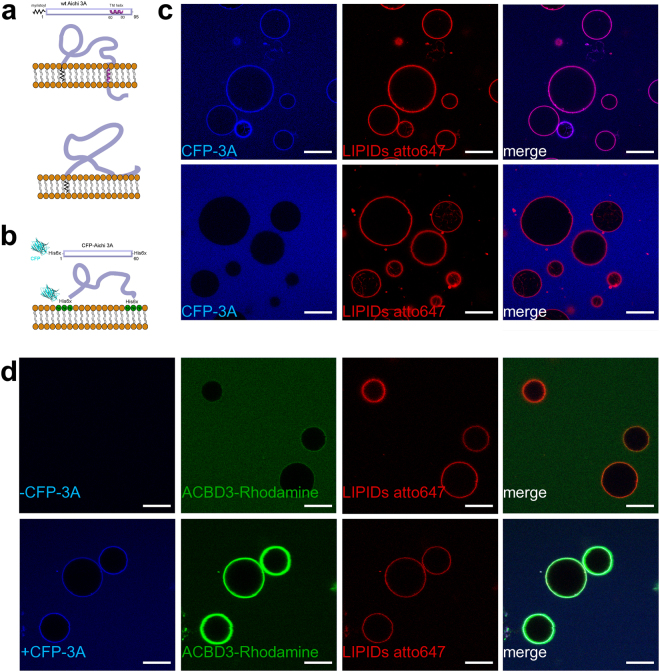
Figure 1.
Aichi virus 3A protein on the membrane. (A) Schematic representation of the wild type 3A protein. The two possible topologies of the wild type 3A protein are shown. Upper panel – the C-terminal hydrophobic stretch is depicted as a transmembrane helix. Lower panel – the C-terminal hydrophobic stretch is depicted as semi-buried in the lipid bilayer. (B) Schematic representation of the mCerulean - 3A fusion protein (named CFP-3A). CFP-3A contains two 6xHis tags; one between the CFP and its N-terminus and one at the C-terminus. These two His tags can be used to attach the CFP-3A protein to a membrane containing DGS-NTA(Ni) (a lipid that has Ni2+ bound to its headgroup). (C) CFP-3A bound to GUVs. Upper panel: 250 nM CFP-3A was added to GUVs containing 5% of DGS-NTA(Ni) and ATTO647N-DOPE (0.1 mol %). Lower panel: as above but DGS-NTA(Ni) was replaced by POPC. The CFP-3A signal is in blue and the ATTO647 signal in red. Representative image of three independent experiments. Scale bar = 20 µm. (D) Aichi virus 3A protein efficiently recruits ACBD3 to the membrane. Rhodamine labeled ACBD3 (250 nM) was incubated with GUVs without 3A protein (upper panel) or with 3A protein (250 nM, lower panel). The CFP-3A signal is in blue, the ACBD3-rhodamine signal is in green, and ATTO647 labeled lipids in in red. Representative image of three independent experiments. Scale bar = 20 µm.