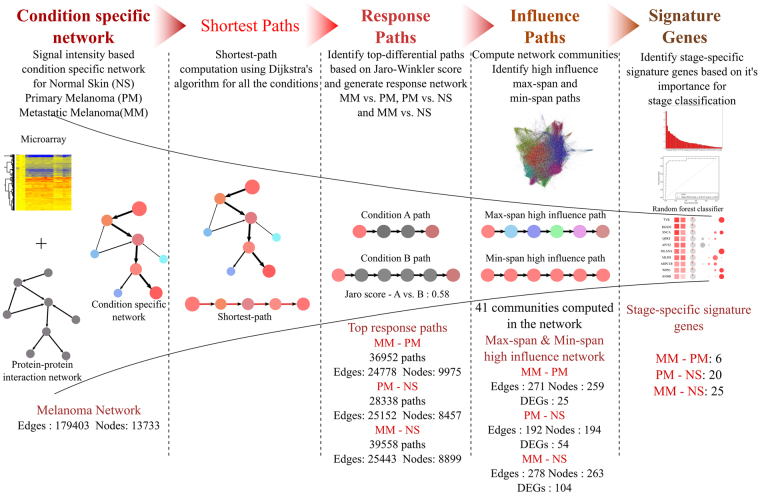
Figure 1.
A schematic representation of the biomarker identification pipeline. The pipeline involves 5 major steps. Condition specific network: Construction of weighted network using protein-protein interaction network and gene expression data. Shortest Path analysis: Identification of all-vs.-all nodes shortest paths using Dijkstra’s algorithm. Response Paths: Paths with highest differential activity in diseased condition identified using string matching metric. Influence paths: Prioritizing paths based on influence on the network. Signature genes: Feature ranking of genes to obtain minimal set classifying conditions. The conditions considered for study: Normal Skin (NS), Primary Melanoma (PM) and Metastasis Melanoma (MM). Analysis results in numbers is shown in bottom section of the image