Figure 5.
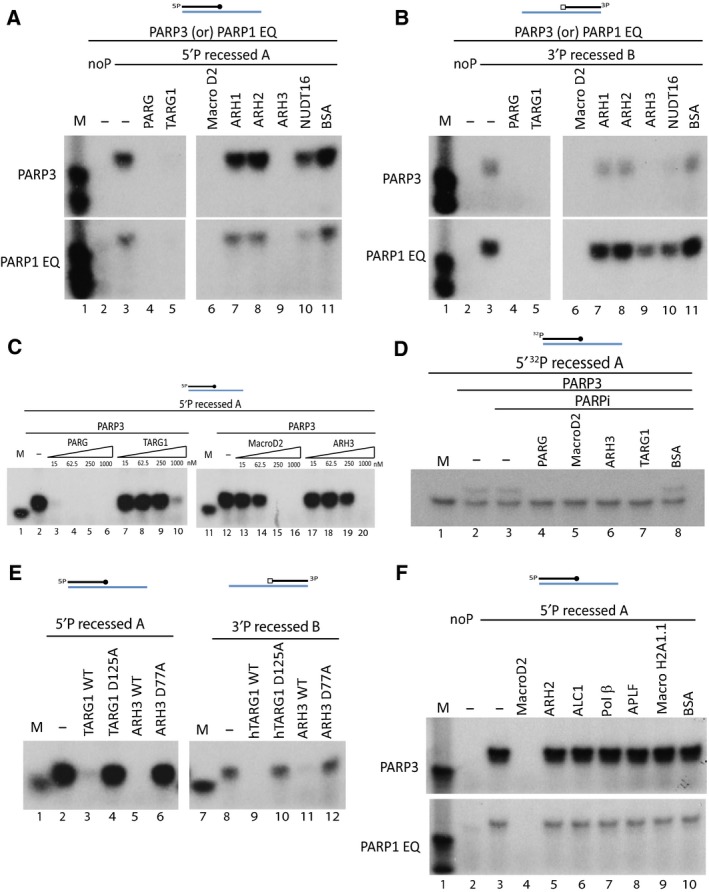
Mono ADP‐ribosylation of DNA is a reversible process. (A) Reversal of ADP‐ribosylated 5′P recessed A substrate by several different hydrolases. (B) ADP‐ribosylated 3′P recessed B substrate is treated with different hydrolases. For both (A) and (B) ADP‐ribosylation of DNA is catalysed by PARP3 (in top panel) or PARP1 E998Q (in bottom panel). (C) PARP3 catalysed ADP‐ribosylation of DNA is subjected to different concentration of hydrolase active in removing DNA ADP‐ribosylation – PARG, TARG1, MacroD2 and ARH3. Hydrolase concentration is titrated from 15 nm to 1000 nm. (D) Mono‐ADP‐ribosylation of 32P labelled DNA substrate is reversed by various hydrolases. (E) Removal of PARP3 mediated ADP‐ribosylation on 5′P recessed A (left panel) and 3′P recessed B (right panel) by wild‐type or point mutant of TARG1 and ARH3. (F) Lack of hydrolytic activity of ADP‐ribosylation‐related proteins – ARH2, ALC1, Polymerase β, APLF and macroH2A1.1 on ADP‐ribosylated DNA substrate catalysed by PARP3 (top panel) and PARP1 E998Q (bottom panel).
