Abstract
Dinoflagellates are prolific producers of polyketide compounds, many of which are potent toxins with adverse impacts on human and marine animal health. To identify polyketide synthase (PKS) genes in the brevetoxin‐producing dinoflagellate, Karenia brevis, we assembled a transcriptome from 595 million Illumina reads, sampled under different growth conditions. The assembly included 125,687 transcripts greater than 300 nt in length, with over half having >100× coverage. We found 121 transcripts encoding Type I ketosynthase (KS) domains, of which 99 encoded single KS domains, while 22 contained multiple KS domains arranged in 1–3 protein modules. Phylogenetic analysis placed all single domain and a majority of multidomain KSs within a monophyletic clade of protist PKSs. In contrast with the highly amplified single‐domain KSs, only eight single‐domain ketoreductase transcripts were found in the assembly, suggesting that they are more evolutionarily conserved. The multidomain PKSs were dominated by trans‐acyltransferase architectures, which were recently shown to be prevalent in other algal protists. Karenia brevis also expressed several hybrid nonribosomal peptide synthetase (NRPS)/PKS sequences, including a burA‐like sequence previously reported in a wide variety of dinoflagellates. This contrasts with a similarly deep transcriptome of Gambierdiscus polynesiensis, which lacked NRPS/PKS other than the burA‐like transcript, and may reflect the presence of amide‐containing polyketides in K. brevis and their absence from G. polynesiensis. In concert with other recent transcriptome analyses, this study provides evidence for both single domain and multidomain PKSs in the synthesis of polyketide compounds in dinoflagellates.
Keywords: algal toxin, dinoflagellate, Karenia brevis, polyketide synthase, toxin biosynthesis
Abbreviations
- ACP
acyl carrier protein
- AT
acyl transferase
- CEGMA
Core Eukaryotic Genes Mapping Approach
- CoA
coenzyme A
- DDBJ
DNA Databank of Japan
- DH
dehydratase
- DNAse
deoxyribonuclease
- DTT
dithiothreitol
- EMBL
European Molecular Biology Laboratory
- ER
enoyl reductase
- FAS
fatty acid synthase
- FeCl3
ferric chloride
- HCS
HMG coA synthetase
- HMG
hydroxymethyl glutarate
- HMM
hidden Markov model
- KR
ketoreductase
- KS
β ketoacyl synthase
- LbH
Left‐handed Parallel beta‐Helix
- MAFFT
Multiple Alignment using Fast Fourier Transform
- MT
methyl transferase
- NRPS
nonribosomal peptide synthase
- P450
cytochrome P450
- Pfam
protein families database
- PhyML
phylogenetics maximum likelihood
- PKS
polyketide synthase
- PP
phosphopantetheine binding site
- RNA
ribonucleotide
- RNAseq
RNA sequencing
- RNAse
ribonucleotidase
- SAM
S‐adenosyl methionine
- TE
thioesterase
- TPP
thiamine pyrophosphate binding domain
- VSSC
voltage sensitive sodium channel
The dinoflagellate Karenia brevis is endemic to the Gulf of Mexico, where it is responsible for red tides that occur almost every year on the west coast of Florida and less frequently in the western Gulf on the Yucatan peninsula and in the northern Gulf from Texas to the Florida panhandle (Steidinger 2009). Karenia brevis blooms have significant environmental impacts, including massive fish kills, marine mammal, bird, and turtle mortalities (Landsberg 2002, Twiner et al. 2012), adverse human health effects through both seafood consumption and respiratory exposure (Fleming et al. 2011), and economic impacts of over $26 million per bloom year (Hoagland et al. 2009, Morgan et al. 2009).
The adverse effects of Karenia brevis blooms are due to their elaboration of a suite of polyether ladder compounds known as brevetoxins, potent neurotoxins that both alter the voltage sensitivity and prolong the open state of voltage sensitive sodium channels (VSSC) involved in both neuronal and neuromuscular synaptic transmission (Baden et al. 2005). Many other dinoflagellate toxins with known health impacts are polyether ladder or related polyketide compounds (e.g., ciguatoxins, diarrhetic shellfish toxins, yessotoxins, amphidinols). Karenia brevis elaborates at least 11 different congeners of brevetoxin bearing two different backbones containing either 10 (A‐type) or 11 (B‐type) trans‐fused ether rings, with variations in side chain constituents that alter their relative toxic potencies (Fig. 1). In addition, K. brevis produces an array of other polyketide compounds (Fig. 1), all with varying affinities for the brevetoxin binding site on the VSSC, including a four‐ring polyether ladder hemibrevetoxin B (Prasad and Shimizu 1989), which binds to but does not activate the VSSC, five‐ring brevenal, which opposes the action of brevetoxins by inhibiting the activation of VSSC (Bourdelais et al. 2004), and a six‐ring “interrupted” polyether ladder, brevesin, whose action is uncharacterized (Satake et al. 2009). In addition, two polyethers contain amide bonds, the seven‐ring tamulamides (Truxall et al. 2010), and a single‐ring compound, brevisamide (Van Wagoner et al. 2010). The extensive elaboration of polyketide compounds by K. brevis and other dinoflagellates suggests they serve an important function. The total cellular concentration of brevetoxins in studied isolates ranges from 1 to 68 pg · cell−1 (Hardison et al. 2012). Based on an estimated 500–700 pg carbon · cell−1 (Kamykowski et al. 1998), with brevetoxins made up of 66% carbon, this equates to an investment of up to 9% of total cellular carbon in brevetoxins alone.
Figure 1.

Diversity of polyketide compounds produced by Karenia brevis.
Stable isotope labeling studies established that brevetoxins are the products of polyketide synthases (PKS; Lee et al. 1986, 1989, Chou and Shimizu 1987). PKSs build carbon chains in a manner analogous to fatty acid synthases (FASs), in which a starting substrate, generally acetyl CoA, is extended through a series of sequential condensations with malonyl CoA, performed by a β‐ketoacyl synthase (KS) domain, to produce a product of predetermined length. In the case of fatty acids, each acetate unit added to the growing chain then undergoes ketoreduction (KR), dehydration (DH), and enoyl reduction (ER), resulting in a fully saturated carbon chain. In contrast, PKSs may lack one or more of these catalytic domains, thereby producing carbon chains that include carbonyl groups (absence of KR domain), hydroxyl (absence of DH domain) or double bonds (absence of ER; Staunton and Weissman 2001). The growing carbon chain resides on a phosphopantetheine “arm” of an acyl carrier protein (ACP) that presents it to the catalytic sites, while an acyl transferase (AT) presents the extender units to be added to the growing chain, most often malonyl coA. The full‐length polyketide is released from the PKS complex by a thioesterase (TE), while post‐PKS modifications create the final polyketide structure. Two major classifications of PKS are found. In Type I PKS (modular) all catalytic domains are found on a single polypeptide, which are used in a progressive fashion for chain elongation, analogous to FASs in animals and fungi (Khosla et al. 1999, Jenke‐Kodama et al. 2005). Type II PKSs are multiprotein complexes where each catalytic domain is found on a separate polypeptide, analogous to type II FASs in bacteria and plants. PKSs typically initiate with an acetyl CoA; however, in dinoflagellates, alternative starter units are sometimes used (Kellman et al. 2010). Furthermore, in brevetoxin, as well as other dinoflagellate polyketides, the carbonyl carbons of some of the acetate units added in the condensation reaction are excised from the growing polyketide by a Favorskii rearrangement (Wright et al. 1996), catalysed by a P450 or flavin monooxygenase. Whereas this step is rare among bacterial and fungal polyketides, it appears to be the norm among dinoflagellate polyketides (Van Wagoner et al. 2014). Another unusual event among dinoflagellate polyketides is the addition of pendent methyl groups from both methionine and acetate. These steps require the involvement of SAM methyl transferase (MT) and HMG‐CoA synthetase (HCS) activity, respectively. Together, these hallmark characteristics of dinoflagellate polyketides suggest that their biosynthetic machinery may include catalytic domains that differ from the better characterized PKSs in bacteria and fungi.
The PKSs of dinoflagellates remain poorly characterized. Sequences with homology to Type I KS domains were first identified in dinoflagellates by Snyder et al. (2005) using degenerate PCR. With the advent of EST libraries, full length transcripts containing Type I‐like KS and KR domains were identified by Monroe and Van Dolah (2008), which were highly unusual in that each contained a single catalytic domain, a 5′ spliced leader sequence and polyA tail, indicating their dinoflagellate origin. These single domain dinoflagellate PKS sequences group phylogenetically with a protist clade, confirming their eukaryotic origins (John et al. 2008). Since that time, single domain PKS sequences have been found in numerous dinoflagellate species (Eichholz et al. 2012, Pawlowiez et al. 2013, Kimura et al. 2015, Kohli et al. 2015) including species considered non‐toxic (Salcedo et al. 2012, Meyer et al. 2015), and single domain Type I‐like PKSs appear to be a characteristic of dinoflagellates. Kohli et al. (2016) mined data from 24 genera and 46 strains of dinoflagellates whose transcriptomes were sequenced under the Moore Foundation's Marine Microbial Eukaryote Transcriptome Sequencing Project, and found that dinoflagellate single domain KSs fall into three clades. Representatives of each clade were present in most dinoflagellates sequenced, and there was no apparent relationship with the evolutionary history of the organism, and no clear relationship with the production of particular known classes of marine biotoxins (Kohli et al. 2016).
The presence of amide‐containing tamulamide and brevisamide (Fig. 1) suggests that Karenia brevis may also encode non‐ribosomal peptide synthases (NRPS) in its genome, which incorporate amino acids in a manner analogous to the incorporation of activated actetate groups by PKS. The first module in an NRPS, known as the initiation module, typically includes an adenylation domain (A) and a peptide carrier protein domain (PCP). Following this are elongation modules which contain a condensation domain (C), A, and PCP domains. Like the ACP domain of PKS, the PCP domain has a post‐translationally added phosphopantetheine arm that passes the peptidyl‐thioester intermediate from one module to the next, with a single amino acid being added at each module. Also analogous to PKS, the full length polypeptide is released by a terminating TE domain. Hybrid NRPS/PKS proteins incorporate both amino acids and acetate building blocks to create a high diversity of natural products. A hybrid NRPS/PKS has previously been reported in K. brevis (Lopez‐Legentil et al. 2010).
With the advent of deeper sequencing afforded by second generation sequencing platforms, there have been several reports of multidomain PKSs present in dinoflagellates (Beedessee et al. 2015, Kohli et al. 2017). Here we assembled a deep transcriptome of K. brevis with a particular interest in mining for the diversity of single domain PKS sequences and the presence of multidomain PKSs, NRPSs, and hybrid NRPS/PKS transcripts.
Methods
Culture conditions
Karenia brevis (nonaxenic, Wilson strain) was grown in sterile filtered seawater at 36‰ and enriched with f/2 media (Guillard et al. 1973) modified with ferric sequestrene in the place of EDTA·Na2 and FeCl3·6H2O and the addition of 0.01 μM selenous acid. All cultures were maintained in 1 L glass bottles at 25°C on a 16:8 h light:dark cycle under cool white light at 45–50 μmol photons · m−2 · s−1. Heat shocked cultures were subjected to 31°C for 30 or 60 min prior to harvesting. This 6°C shock results in a significant decrease in translation, as evidenced by the loss of polysomes, a classical hallmark of stress (Fridey 2015).
RNA extraction
Whole cultures were harvested by centrifugation at 600g. RNA extraction was performed on the pelleted cells using TriReagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer's protocol. Extracted RNA was DNase treated with RQ1 RNase‐free DNase (Promega, Madison, WI, USA) for 10 min at room temperature and the RNA was then purified on columns using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) following the manufacture's protocol.
To obtain actively translated RNA pools, 50 mg · mL−1 cycloheximide was added to each L of K. brevis and incubated for 5 min to prevent ribosome runoff prior to harvesting. The cells were then harvested by centrifugation at 600g and pellets homogenized in polysome resuspension buffer (50 mM Tris HCL, pH 8, 100 mM NH4Cl, 20 mM sucrose, 10.5 mM Mg acetate, 0.5 mM EDTA, 1 mM DTT, 0.1% heparin, 0.01% cycloheximide, 0.1% Triton X‐100, 80 U RNAsin) and centrifuged at 15,000g for 10 min. Supernatants were layered on 12.5%–50% sucrose gradients, which were centrifuged at 180,000 × g at 4°C for 2.5 h in a Beckman SW 41Ti rotor. Polysomes were then profiled by pumping the sucrose gradients through a flow cell with UV detection at 254 nm (ISCO). One milli liter sucrose fractions representing the polysome fractions (2–6 ribosomes) were pooled and RNA extracted by the addition of 1 part 8 M guanidine HCL. One part ethanol was then added and samples were incubated at −80°C overnight; then 2 μL of polyacryl carrier was added to facilitate RNA precipitation. Samples were centrifuged at 12,000g for 30 min at 4°C, washed with cold 75% ethanol, and resuspended in nuclease‐ free water with RNasin Ribonuclease Inhibitor (Promega), and DNase treated as described above. All RNA samples were quantified using a NanoDrop N‐1000 Spectrophotometer (Thermo Scientific, Woltham, MA, USA) and integrity assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
RNAseq libraries
A total of 595 million Karenia brevis reads were assembled into one transcriptome. The constituent reads included several RNAseq libraries, generated under different conditions and using different library generation protocols and sequencing platforms. The first two libraries were generated from cells harvested at mid‐log phase (MMETSP201, 56 M reads) or stationary phase (MMETSP202, 44 M reads) and submitted to the Moore Foundation Marine Microbial Eukaryote Transcriptome Sequencing Project for library preparation using Illumina Trueseq RNA protocol. These libraries generated ~25 million 50 nt paired‐end sequencing reads on Illumina HiSeq2000. These sequences, available from NCBI Short Read Archive number SRR042159, were pooled to yield 100 million reads. Nine stranded libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit (Illumina, Hayward, CA, USA) from total RNA of log phase cultures under control, exponential growth conditions (n = 3), or cultures exposed to 30 min (n = 3) or 60 min (n = 3) of 6°C heat shock prior to harvest as described in Fridey (2015). Sequencing of these libraries was performed on an Illumina Hiseq 2500 sequencer, at a depth of ~25 million, 125 nt, single end reads per library. Pooling of these libraries resulted in 225 million single end reads. Lastly, nine libraries consisted of RNA isolated from the translationally active RNA pools isolated from polyribosomes as described by Fridey (2015) from log phase K. brevis cells under control exponential growth conditions (n = 3), or cultures exposed to 30 min (n = 3) or 60 min (n = 3) of 6°C heat shock prior to harvest. These libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina) and on Illumina HiSeq2000 at a depth of ~14 million 50 nt paired‐end reads. These sequences were pooled to yield 270 million reads. These sequences are available from NCBI Bioproject number PRJNA343279.
Transcriptome assembly and analysis
Raw reads from individual libraries were quality filtered, and then all reads were assembled together into contigs using CLC Genomics Workbench (CLC Bio, Cambridge, MA, USA) with default settings. Contigs with a length of less than 300 nt were not analyzed further. The combined transcriptome has been deposited in DDBJ/EMBL/GenBank under the accession GFLM01000000. BLASTx analysis, mapping, annotation and Kyoto Encyclopedia of Gene and Genomes analysis for both the gene catalogues was performed using Blast2GO (Conesa et al. 2005). BLASTx was performed against the nr database of GenBank using an E‐value cut‐off of 10−3. For mapping and annotations, the default values of Blast2GO were used. To analyze the comprehensiveness of the gene catalogues the Core Eukaryotic Genes Mapping Approach (CEGMA) tool was used (Parra et al. 2007). HMMER (Finn et al. 2011a, b) was used for the identification of contigs containing conserved PKS domains (KS, KR, ACP, AT, DH, ER, TE) using an in‐house HMM database (Kohli et al. 2017). Functional prediction of sequences was further analyzed by Pfam (Punta et al. 2012) and conserved domain searches (Marchler‐Bauer et al. 2014) were used for identification of conserved amino acid residues and functional prediction of PKS and PKS/NRPS transcripts. Phylogenetic analysis steps were performed in Geneious software (Kearse et al. 2012). Sequences were aligned using MAFFT v6.814b (Katoh et al. 2002) and/or ClustalW (Thompson et al. 1994). Alignments were trimmed manually to ensure they spanned the KS or KR conserved domain. Maximum likelihood phylogenetic analysis was carried out using PhyML (Guindon et al. 2010) or RAxML Version 7.0 (Stamatakis 2006) using LG model of rate heterogeneity with 1,000 bootstraps. Phylogenetic trees were visualised using MEGA:Version 6 (Tamura et al. 2013).
Results and Discussion
A transcriptome of Karenia brevis assembled from 595 million Illumina reads, pooled from libraries representing different growth stages (log and stationary phase) and stress conditions (heat shock), produced 125,687 contigs greater than 300 nt in length. The assembly had an N50 of 1,829 nt and an average contig length of 1,413 nt, where the longest contig generated was 36,109 nt and over 400 contigs were >8,000 nt in length. The GC content was 52.4%, consistent with previous K. brevis libraries reported (Lidie et al. 2005, Ryan et al. 2014), similar to Symbiodinium, which varies by isolate (50.5%–56.4%; Bayer et al. 2012), and lower than those of other peridinin containing dinoflagellates (~60%; Jaeckisch et al. 2011, Kohli et al. 2017). Over half (53.6%) of the contigs had >100× coverage, supporting their assembly (Table 1). Overall, 55.1% of the contigs had BLASTx hits to the Genbank non‐redundant database (E‐value <10−3), of which 54.4% had annotated matches, while 45.6% matched uncharacterized proteins. The percentage of annotated matches observed increased with increasing contig length and coverage (Table 1).
Table 1.
Coverage and annotation statistics of Karenia brevis gene catalogue. An e‐value cut‐off of 10−3 was applied during BLASTx analysis
| Coverage | No. of contigs | Length (mean) | BLASTx analysis | PKS sequences | ||
|---|---|---|---|---|---|---|
| Annotated match | Non‐annotated match | No match | ||||
| 1×–5.00× | 2,819 | 300–1,401 (409.3) | 167 | 310 | 2,342 | 7 |
| 5.01×–20× | 15,033 | 300–10,974 (817.5) | 2,455 | 3,046 | 9,532 | 14 |
| 20.01×–50× | 19,906 | 300–24,286 (1,292.2) | 5,278 | 5,425 | 9,203 | 5 |
| 50.01×–100.00× | 20,907 | 300–36,109 (1,536.5) | 6,804 | 5,396 | 8,707 | 10 |
| 100.01×–1,000× | 61,676 | 300–34,047 (1,641.8) | 20,692 | 16,265 | 24,719 | 91 |
| 1,000.00×–10,000× | 5,272 | 300–6,989 (956.6) | 2,291 | 11,09 | 1,872 | – |
| 10,000.01× and above | 74 | 311–2,254 (543.9) | 58 | 5 | 11 | – |
| Total number of contigs (percentage) | 125,687 | 300–36,109 (1,413.3) | 37,745 (30%) | 31,556 (25.1%) | 56,386 (44.9%) | 127 |
To further assess the completeness of the transcriptome assembly, we conducted an analysis for the presence of 248 ultra‐conserved core eukaryotic genes using CEGMA. We found 223 (88%) of the 248 highly conserved genes present in full length, with 90% recorded as present if partial matches are included. This is higher than previously reported transcriptome assemblies of K. brevis (Ryan et al. 2014) and likely reflects the greater depth of sequencing (595M reads) included in the current transcriptome. A description of sequences encoding essential enzymes for fifteen conserved metabolic pathways is presented in Table S1 in the Supporting Information.
Polyketide synthases
Using HMMER and conserved domain searches to identify ketosynthase domains, we found 121 KS containing contigs: 99 possess a single KS domain while 22 contain multiple KS domains (Table S2 in the Supporting Information). A phylogenetic analysis of KS domains from K. brevis and those reported from Gambierdiscus spp. (Kohli et al. 2017) placed all dinoflagellate single‐KS domain sequences, and most dinoflagellate KS domains found in multidomain transcripts, within a clade made up of KS sequences from other protists (Fig. 2). This provides evidence that both the single domain transcripts and multidomain KS sequences are of dinoflagellate origin, and not due to bacterial or fungal contaminants known to co‐occur in K. brevis cultures. Further evidence for the dinoflagellate origin of the single domain sequences is the presence of the dinoflagellate‐specific spliced leader sequence on the 5′ ends of at least 12 transcripts containing the full 5′ end. Within the protist clade, the single domain K. brevis KSs form a monophyletic sub‐branch, while multidomain KS sequences formed several independent clades that grouped with the Apicomplexa. A small subgroup of multidomain dinoflagellate KSs fell within a bacterial clade as discussed below.
Figure 2.
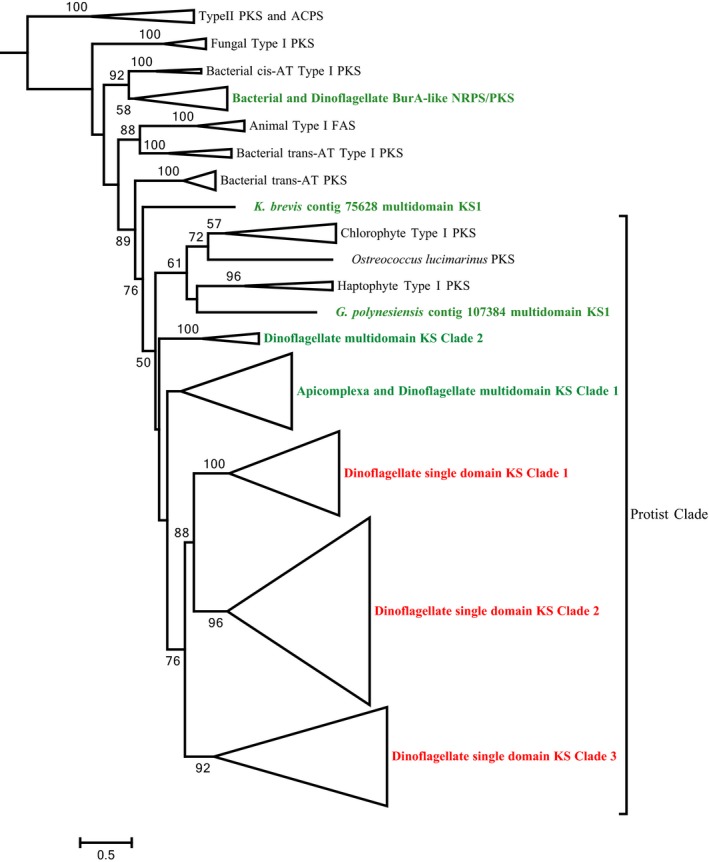
Phylogenetic analysis of ketoacyl synthase (KS) domains from prokaryotic and eukaryotic Type I and II polyketide synthases (PKS) and Type I FAS. The alignment included 121 KS domains from Karenia brevis sequences encoding single and multidomain PKS and NRPS/PKSs and 154 KS domains from Gambierdscus polynesiensis and other prokaryotic and eukaryotic taxa. Analysis was conducted using RAxML, using the LG model of rate heterogeneity, with 1,000 bootstraps. Bootstrap values ≥50% shown.
Single domain KS sequences
Within the dinoflagellate single‐domain KS clade, three sub‐clades exist with good bootstrap support (Fig. 2), here termed Clades 1, 2, and 3. These clades appear to correspond with three single‐domain KS clades identified by Kohli et al. (2016) in a survey of dinoflagellate transcriptomes. Single domain KS sequences identified in Karenia mikimotoi similarly fall into three clades (two clades, one with two major subclades; Kimura et al. 2015) with the same K. brevis membership as described below.
Clade 1 includes previously published Karenia brevis KS proteins KB2006 and KB5361 (Monroe et al. 2008). All K. brevis sequences within this clade have the conserved active site cysteine (C) required for decarboxylative condensation, within the conserved sequence (D/N)TACS(S/A)(S/G) sequence, originally identified as DTACSSS in the crystal structure of Escherichia coli FabH (Davies et al. 2000; Fig. 3). All members of this clade also have the conserved histidine (H) within the conserved HGTGT sequence required for transacylation (Davies et al. 2000). The second H required for KS activity is present in all sequences with a consensus of NIGH. Although the tree in Figure 2 was generated using only the KS domain, when a multiple alignment is carried out using the full‐length sequences in this clade, all but three sequences have the conserved GYLG motif reported previously in the 5′ region of single domain dinoflagellate KSs (Eichholz et al. 2012, Pawlowiez et al. 2014).
Figure 3.
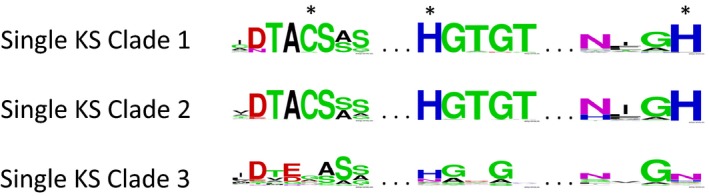
Consensus sequences found in the conserved active sites within single domain KS sequences. Sequences in Clades 1 and 2 possess the highly conserved cysteine and histidines (starred), whereas sequences in Clade 3 are divergent, most lacking the conserved C and Hs. Consensus sequences were generated using Weblogo (http://weblogo.berkeley.edu/logo.cgi).
Clade 2 Karenia brevis KS sequences in this clade all have the conserved active site C required for decarboxylative condensation, within the sequence (D/E)TACS(S/T/A)(S/A/G/M; Fig. 3). All members of this clade have the conserved H within H(G/A/C)TGT sequence required for transacylation. The second conserved H required for KS activity is also present in all sequences in this clade, with a consensus of NIGH. Sequences in this clade contained a consensus of AYLG in the 5′ dinoflagellate motif, with variations in the first two positions quite frequent. This clade included the KB6736 previously described by Monroe and Van Dolah (2008).
Clade 3 included 16 Karenia brevis sequences with highly divergent active sites, including transcripts identical to previously reported KS sequences KB1008, KB4361, and KB6842. No sequences in this clade possess an active site C (in the expected sequence DTACSS) required for decarboxylative condensation. In its place is a consensus of DXEX(A/S)S (where X is too variable to obtain a consensus; Fig. 3). The conserved H in the expected HGTGT is present in only about half of the sequences in this clade, with a consensus sequence of HGXGX. The second conserved H is replaced in most sequences in this clade with N, with a consensus of NXGN. Given these differences in the active sites that are conserved in all known KSs, it is unclear what function(s) these sequences may have. When the full‐length sequences are aligned, the conserved 5′ dinoflagellate motif is present as GLLG, the original sequence reported by Eichholz et al. (2012) and observed also in Gambierdiscus polynesiensis (Pawlowiez et al. 2014).
Single domain PKS KS sequences lack plastid targeting peptides
Insight into the presence of organelle targeting sequences on transcripts can be informative for discerning the metabolic processes they are involved in. To this end, we analyzed the 5′ ends of all full length single domain K. brevis KS sequences for the presence of signal peptides using SignalP predictor (http://www.cbs.dtu.dk/services/SignalP/), beginning with the translation initiating methionine. Signal peptides were absent from all PKS KS domains. In contrast, signal peptides were present on the six Type II FAS KS domain transcripts (KASII and III oxoacyl ACP synthases) present in the K. brevis transcriptome (Table 2). Previous analyses show that dinoflagellate FASs reflect the phylogenetic history of their plastids and are predicted to be plastid localized based on the presence of targeting sequences (Kohli et al. 2016). Karenia brevis possesses a tertiary plastid of haptophyte origin, and its chloroplast transit peptides differ from those in peridinin dinoflagellates, in which the signal peptide cleavage site is typically followed by a conserved sequence FVAP (Patron et al. 2006). The signal cleavage sites in K. brevis KAS II sequences and several known plastid targeted proteins examined (Table 2) are preceded by G and followed by RRV(K/Q), while the sequence in KASIII is G‐RRIL. A similar conserved sequence RRxQ was found in 26 plastid targets proteins in another tertiary plastid containing dinoflagellate, Karlodinium micrum (Patron et al. 2006). The absence of such plastid targeting sequences on single domain PKS KS transcripts would predict cytosolic localization. Confoundingly, the only single domain KS protein to be studied at the protein level, KB2006 (Monroe et al. 2010) was localized to the chloroplast pyrenoid by immunolocalization and to the chloroplast by subcellular fractionation (Monroe et al. 2013, Van Dolah et al. ). If the PKS biosynthetic machinery is in fact located in the chloroplast, it is unclear what mechanism transports these proteins to the plastid.
Table 2.
Predicted signal peptides and cleavage sites in known plastid‐localized proteins and Type II FAS sequences in Karenia brevis. Signal peptide prediction and score generated by signalP. The D‐score (discrimination score) is used to discriminate signal peptides from non‐signal peptides, using a threshold of 0.5. Cleavage site sequences were screened for the conserved FVAP sequence found in peridinin dinoflagellates. In some sequences a plausible FVAP‐like sequences is present, but most possess the RRV(Q/K) also present in Karlodinium. No signal peptides were present in any single domain PKS KS or KR sequences
| GenBank accession no. or contig no. | CD and blast ID | Prediction | D score | Cleavage site |
|---|---|---|---|---|
| ABF73013 | Cytochrome b6f FeS subunit | No | 0.131 | – |
| Contig 15936 | Cytochrome b6‐f | Yes | 0.55 | G‐QSPREQ |
| ABF73015 | Ferredoxin | Yes | 0.613 | R‐RRVRD |
| Contig 15936 | Multimeric flavodoxin | Yes | 0.763 | A‐ASLAS |
| ABF73016 | Flavodoxin nadph reductase | Yes | 0.673 | G‐FRVQ |
| ABF73017 | Oxygen enhancer | No | 0.419 | G‐RFQQK |
| ABF73018 | psII 12kD | Yes | 0.864 | A‐FSPA |
| ABF73029 | GapDH C1 | Yes | 0.606 | A‐(+9)FEEQ |
| ABF73002 | GapDH C1 | No | 0.323 | A‐FIAPA |
| kbrevis_combined_contig_386 | KASII 3 oxoacyl ACP synthase | Yes | 0.7 | G‐RRVQ or G‐(+9)FKPA |
| kbrevis_combined_contig_387 | KASII 3 oxoacyl ACP synthase | Yes | 0.681 | G‐RRVQ or G‐(+9)FKPA |
| kbrevis_combined_contig_46229 | KASII 3 oxoacyl ACP synthase | Yes | 0.723 | S‐DYGR or G‐RRVK |
| kbrevis_combined_contig_37304 | KASII 3 oxoacyl ACP synthase | Yes | 0.607 | G‐RRVQ or A‐FNPA or G‐(+9)FNPA |
| kbrevis_combined_contig_49622 | KASIII 3 oxoacyl ACPsynthase | Yes | 0.816 | G‐RRIL |
| kbrevis_combined_contig_275 | KASII 3 oxoacyl ACP synthase | Yes | 0.572 | G‐RRLN |
| kbrevis_combined_contig_97889 | KR | Yes | 0.837 | A‐QTPT or A‐(+9)FPHA |
| kbrevis_combined_contig_64411 | KR | Yes | 0.479 | G‐RPMQ |
| kbrevis_combined_contig_86499 | KR | Yes | 0.798 | A‐YGEF or C‐SSRE |
| kbrevis_combined_contig_28593 | ER | Yes | 0.646 | G‐KRVQ |
| kbrevis_combined_contig_47965 | ER | Yes | 0.69 | G‐KRVK |
| kbrevis_combined_contig_44484 | DH | No | 0.148 | – |
| kbrevis_combined_contig_6314 | AT | Yes | 0.659 | G‐RTLQ |
| kbrevis_combined_contig_101182 | AT | No | 0.427 | G‐RRLQ |
Multidomain KS containing transcripts
Twenty‐four contigs were identified as multidomain PKS (16), NRPS/PKS (6), or NRPS (2) encoding one to three modules (Table 3). Both cis‐ and trans‐AT architectures were present. None of the multidomain sequences includes a 5′ SL sequence, suggesting that they may not be complete, or they may not be processed via SL trans‐splicing.
Table 3.
Domain structure of multidomain PKS, NRPS/PKS hybrids, and NRPS contigs in the Karenia brevis assembly
| Contig number | Length (nt) | Classification | Cis/Trans AT | No. of modules | Domain structure |
|---|---|---|---|---|---|
| Contig 1930 | 19,025 | NRPS/PKS | trans | 3 | TE‐A‐DH‐PP‐C‐A‐PP‐KS‐DH‐KR‐PP‐KS‐DH‐KR‐PP‐PP‐PP‐PP‐PP‐PP‐TE‐(MT) |
| Contig 10563 | 18,781 | NRPS/PKS | trans | 3 | TE‐A‐DH‐PP‐C‐A‐PP‐KS‐DH‐KR‐PP‐KS‐DH‐KR‐PP‐PP‐PP‐PP‐PP‐PP‐TE‐(MT) |
| Contig 5155 | 11,047 | PKS | trans | 2 | PP‐KS‐DH‐KR‐PP‐KS‐DH‐KR‐PP‐PP‐PP‐PP‐PP‐PP‐TE‐(MT) |
| Contig 78360 | 2,221 | PKS | trans | 3 | PP‐KS‐KR‐PP‐KS‐KR‐PP‐KS‐KR‐PP‐TE‐PP‐LbH |
| Contig 10709 | 14,778 | PKS | trans | 3 | PP‐KS‐DH‐KR‐PP‐KS‐KR‐PP‐KS‐DH‐KR[ER]KR‐TE |
| Contig 15957 | 3,031 | PKS | trans | 1 | KR‐PP‐KS‐KR‐PP‐TE‐TE‐LbH |
| Contig 81604 | 7,799 | PKS | trans | 3 | (KS)‐KR‐PP‐KS‐KR‐PP‐KS‐KR |
| Contig 28414 | 12,319 | PKS | cis/trans | 2 | PP‐KS‐AT‐DH‐KR[ER]KR‐PP‐KS‐KR |
| Contig 54805 | 8,035 | PKS | cis | 1 | KS‐AT‐KR‐LbH‐PP‐PP‐LbH‐PP‐PP‐LbH |
| Contig 75628 | 10,131 | PKS | cis | 1 | KS‐AT‐DH‐KR‐PP‐KS‐(AT)‐KR‐TE |
| Contig 57200 | 3,254 | PKS | ? | 1 | PP‐KS‐DH |
| Contig 89014 | 1,027 | PKS | ? | 1 | PP‐KS |
| Contig 99638 | 3,763 | PKS | ? | 1 | PP‐KS‐KR‐PP |
| Contig 113789 | 2,654 | PKS | ? | 1 | (KS)‐KR |
| Contig 114143 | 2,119 | PKS | ? | 1 | PP‐KS |
| Contig 124885 | 3,014 | PKS | ? | 1 | KS‐KR |
| Contig 134145 | 2,544 | PKS | ? | 1 | PP‐KS |
| Contig 34829 | 18,811 | NRPS/PKS | trans | 2 | tpp‐A‐KR‐PP‐KS‐PP‐C‐TE‐A‐PP‐C‐A‐PP‐TE |
| Contig 10632 | 9,386 | NRPS/PKS | cis | 1 | A‐KS‐AT‐DH‐ER‐PP‐(TE) |
| Contig 3318 | 8,238 | NRPS/PKS | cis | 1 | TE‐A‐PP‐KS‐AT‐TE‐KR‐PP |
| Contig 77766 | 4,288 | NRPS | ? | 2 | PP‐C‐A‐PP‐(C) |
| Contig 4898 | 2,638 | NRPS | ? | 2 | TE‐A‐DH‐PP‐C‐A |
KS, ketosynthase; KR, ketoreductase; DH, dehydratase; ER, enoyl reductase; KR(ER)K, KR with embedded ER domain; AT, acyl transferase; TE, thioesterase; A, adenylation domain; C, condensation domain; PP, phosphopantetheine binding site of ACP or PCP domains; (MT), possible methyl transferase; tpp, thiamine pyrophosphate binding omain; LbH, left‐handed beta helix; ( ), partial domain; [ER], embedded within KR domain.
Trans‐AT PKSs
A majority of the Karenia brevis multidomain PKSs lack AT domains (Table 3). Trans‐AT PKSs were first identified over a decade ago in bacteria, and since been found broadly among bacteria and protists. Trans‐AT PKSs evolved independently of the better known cis‐AT PKSs, both originating from FASs (Piel 2010). Whereas cis‐AT PKSs have only eight possible domain architectures (KS‐AT‐ACP, KS‐AT‐KR‐ACP, KS‐AT‐DH‐KR‐ACP, KS‐AT‐DH‐ER‐KR‐ACP, with and without MT), there are more than 50 module types known to date in trans‐AT PKSs (Piel 2010). Trans‐AT PKSs are distinguished from cis‐ATs not only in the lack of AT domains, but also unusual ordering of domains, inclusion of unusual domains, the presence of non‐condensing KS0 domains, and the amplification of ACP domains. Trans‐AT PKSs have been found to be common among other microalgal PKSs (Shelest et al. 2015). In the phylogenetic analysis of K. brevis KS domains, most of the multidomain KSs fall within a clade that includes apicomplexan PKSs (Fig. 2; Dinoflagellate multidomain KS Clade 1). All K. brevis KS domains in this clade are from trans‐AT sequences or trans‐AT modules within mixed cis/trans‐AT sequences from K. brevis and G. polynesiensis (Fig. 4A).
Figure 4.
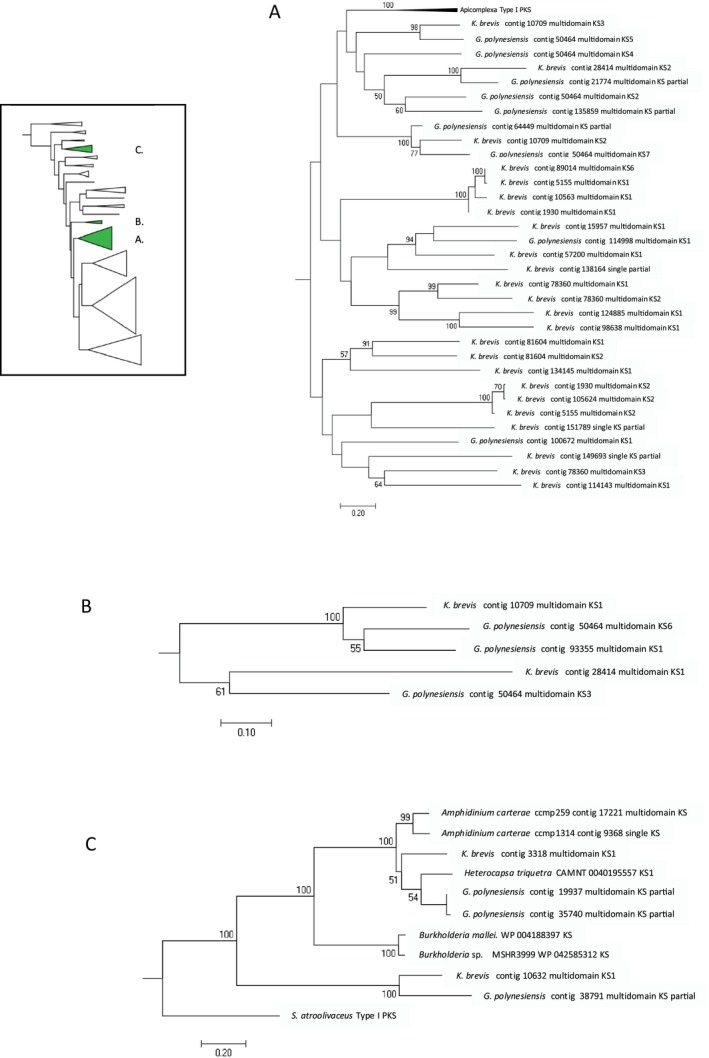
Details of clade membership of Karenia brevis KS domains in Figure 2. Multidomain PKS sequences were found primarily in three clades. Inset shows locations of clades detailed in this figure. (A) Dinoflagellate multidomain Clade 1 within the protist clade that included apicomplexans. This clade contains KS domains from K. brevis and Gambierdscus polynesiensis trans‐AT PKSs. (B) Dinoflagellate multidomain Clade 2 within the protist clade includes two cis‐AT and one trans‐AT PKS in K. brevis and several trans‐AT G. polynesiensis KSs. (C) Clade 3 includes bacterial and dinoflagellate BurA‐like NRPS/PKS, cis‐AT NRPS/PKS sequences and cis‐AT bacterial Type I PKS. S. atroolivaceus, Streptococcus atroolivaceus.
Cis‐AT PKSs
Two PKS transcripts encode cis‐AT domains. Contig 75628 falls within the protist clade but is outside of the dinoflagellate KS clades. Contig 28414 has one cis‐ and one trans‐AT module (ACP‐KS‐AT‐DH‐ER‐KR‐ACP‐KS‐KR). The trans‐AT module falls within the dinoflagellate trans‐AT clade (Fig. 2 multidomain Clade 1 and Fig. 4A), while the cis‐AT module falls within Dinoflagellate multidomain KS Clade 2 (Figs. 2 and 4B), along with K. brevis contig 10709 and several G. polynesiensis KS sequences, which appear to be from trans‐AT PKSs. Thus, the cis‐AT associated KSs from PKS sequences did not form a unique clade.
NRPS/PKSs
Six hybrid NRPS/PKS sequences were identified in the Karenia brevis assembly. Sequences with identical domain architecture to Contig 3318 (TE‐A‐PP‐KS‐AT‐TE‐KR‐PP) have been reported previously in a wide array of dinoflagellates, from the basal dinoflagellate Oxyrrhis to 18 core dinoflagellates, including K. brevis (Bachvaroff et al. 2014), as well as in Gambierdiscus spp. (Kohli et al. 2017). This domain architecture is found in the burA gene from the bacteria Burkholderia. BurA is involved in the synthesis of one of two polyketide precursors of burkholderic acid, an unprecedented furan derivative produced from the fusion of two polyketide chains (Franke et al. 2012). The transcript has both an N‐terminal TE domain and an internal TE. The function of an N‐terminal TE domain is not yet understood. Bachvaroff et al. (2014) reported a dinoflagellate spliced leader on the 5′ end of this transcript in Amphidinium carterae, suggesting its dinoflagellate origin, although the SL was supported by a single overlapping read. In our phylogenetic analysis, the KS domain from Contig 3318 falls in a clade with other dinoflagellate KS domains from cis‐AT NRPS/PKS sequences and bacterial burA (Figs. 2 and 4C).
NRPS/PKS Contigs 1930 and 10563 encode an N‐terminal NRPS module, followed by two trans‐AT PKS modules that group with other trans‐AT KS domains within the protist clade. The second PKS module in both sequences has six tandem ACP domains, followed by a potential MT (M) domain of the SAM dependent methyltransferase superfamily. Duplicated ACP domains are not unusual in trans‐AT PKSs, although six tandem repeats have not previously been reported. In bacterial trans‐AT PKSs, the presence of repeated ACP domains typically follows KS domains that lack the capacity for condensation and it has been proposed that their presence provides longer dwell time for substrates (Piel 2010). However, the KS domains in the K. brevis NRPS/PKSs preceding the tandemly repeated ACPs appear to be fully functional based on the presence of the conserved active site C and Hs. Contigs 1930 and 10563 are 91% similar at the amino acid level, with differences primarily occurring as short stretches of amino acid inserts. Contig 5155 lacks their N‐terminal NRPS module, but is 90% similar to these sequences in their PKS domains. The first KS domain in these sequences has 91% similarity to AT2‐10L, a KS domain identified by Snyder et al. (2003) by degenerate PCR and shown to be expressed in the dinoflagellate (and not co‐occurring bacteria) by fluorescence in situ hybridization (Snyder et al. 2005). AT2‐10L has not previously been assigned to a multidomain PKS/NRPS, but was shown to cluster separately from single domain KS domains (Monroe and Van Dolah 2008). These sequences are also unusual in that, like Contig 3318 described above, they possess a TE domain at the N‐terminal end.
Comparison with multidomain PKS identified in other dinoflagellates
To date few publications describe multidomain PKSs in dinoflagellates. Bachvaroff et al. (2014) provided the first report of a multidomain PKS/NRPS hybrid in a large array of dinoflagellates with architecture similar to the burA gene in Burkholderia as described above. Kohli et al. (2017) conducted a survey of PKSs in four species of Gambierdiscus, sequenced at depths between 134 million to 1 billion Illumina reads, and assembled in CLC Workbench using similar parameters as used here for K. brevis. In contrast to K. brevis, where a quarter of the sequences recovered were NRPS/PKS hybrids, only the deepest library (G. polynesiensis) revealed any NRPS/PKS sequences, a contig similar to the burA‐like sequence discussed above and a short contig containing domains C‐ACP‐KS. Unlike K. brevis, which produces at least two amide containing compounds (talulamide and brevisamide; Fig. 1), no products of NRPS pathways are known in Gambierdiscus spp. Like K. brevis, both cis‐ and trans‐AT PKS sequences were present in Gambierdiscus spp. The longest sequence found in G. polynesiensis was a 7‐module PKS, of which the first six modules lacked AT domains while the seventh encoded a cis‐AT. The predicted product of this sequence resembles the carbon backbone of a polyether ladder compound. No sequence of this length this was found in K. brevis. However, the KS domains in this sequence cluster with trans‐AT sequences in K. brevis that have similar domain architectures. All other contigs found in Gambierdiscus encoded only one module or partial modules, whereas in K. brevis, half of the PKS or NRPS/PKS contigs contained 2–3 modules. Also absent from the Gambierdiscus assemblies were the tandemly repeated ACP domains observed in some K. brevis NRPS/PKS and PKS sequences. Beedessee et al. (2015) reported 10 multidomain PKS and NRPS/PKS hybrid genes in a genomic survey of Symbiodinium minutum, with transcriptome support indicating that these genes are expressed as the predicted multidomain sequences. Both cis‐ and trans‐AT PKS sequences were present. One sequence contained a singly repeated ACP domain but none contained highly repeated ACPs as found in K. brevis. One very large 8‐module NRPS/PKS hybrid sequence of 10,601 amino acids was found. Most of the PKSs reported encoded single modules (7), 2 modules (1) or 3 modules (1). In some cases, multiple gene models with transcript support were found on the same scaffold, suggesting that the smallish (1–3 module) transcripts that have been identified in K. brevis and other dinoflagellate transcriptomes may represent full transcripts and not partial assemblies of larger multimodular transcripts. Confirmation of their completeness may require genomic sequence data for K. brevis.
KR sequences
Using HMMER and conserved domain searches to identify ketoreductase domains, we found 8 transcripts encoding single KR domains, while 25 were present on the multidomain PKS sequences described above (Table 2; Table S2). All K. brevis KR domains possess the conserved active site residues YXXXN present in Type I PKS and animal FAS that distinguishes them from KR domains in Type II FAS/PKS, which possess YXXXK. The number of unique single domain KRs is much smaller than the number of single domain KS sequences described above (n = 95), suggesting that their selection is more highly conserved. A similar trend was observed in four Gambierdiscus spp. (expressing collectively 90 single domain KS and 7 single domain KR domains; Kohli et al. 2017). Phylogenetic analysis placed all K. brevis KR domains within a clade that included protist ‐ chlorophyte, haptophyte, and Apicomplexa ‐ and bacterial KR domains (Fig. 3). Within this clade, the single domain KR sequences formed a monophyletic group separate from the KR domains found in multidomain PKSs (Fig. 4). The KR domains from multidomain PKSs formed a clade with several interesting subclades (Figs. 5 and 6). Sub‐clade 1 is made up entirely of KR domains from PKS modules with the architecture KS‐KR‐ACP. Sub‐clade 2 is made up primarily of KR domains from trans‐AT PKS modules with the architecture KS‐DH‐KR. Sub‐clade 3 KR domains possess an ER domain inserted between the N‐terminal and C‐terminal subdomains of the KR. This architecture has been observed previously in porcine FAS (cd05275; https://www.ncbi.nlm.nih.gov/cdd/). Subclades 4 and 5 include all NRPK/PKS sequences found in K. brevis. Clade 4 includes burA‐like sequences in K. brevis and other dinoflagellates as well bacterial burA genes from Burkholderia spp. Clade 5 includes three highly similar contigs in K. brevis with module architecture KS‐DH‐KR ‐ two NRPS/PKS (contig 1930, contig 10563) ‐ and one PKS (contig 5155) that lacks the N‐terminal NRPS module (Table 2). These sequences have in common highly amplified ACP domains (n = 6) and potential MT domains at their c‐terminal ends. In general, KR domains clustered according to their module architecture rather than grouping with other KR domains in a given multimodule sequence, if their module architectures differed.
Figure 5.
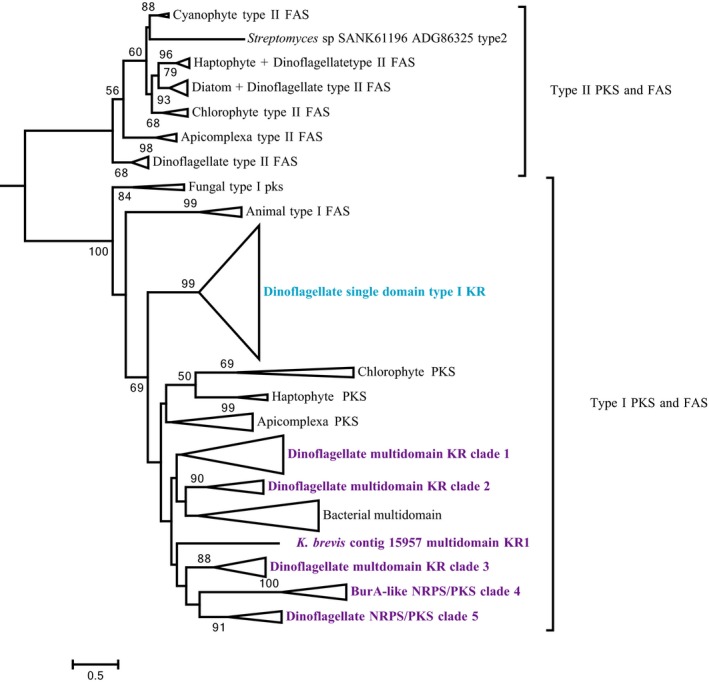
Phylogenetic analysis of ketoacyl reductase (KR) domains from prokaryotic and eukaryotic Type I and II PKS and Type I and II FAS. The alignment included 193 KR domain sequences, including 33 encoding single and multidomain PKS and NRPS/PKSs from Karenia brevis, and 160 from other dinoflagellates and other prokaryotic and eukaryotic taxa. Analysis was conducted using RAxML using the LG model of rate heterogeneity, with 1,000 bootstraps. Bootstrap values ≥50% shown.
Figure 6.
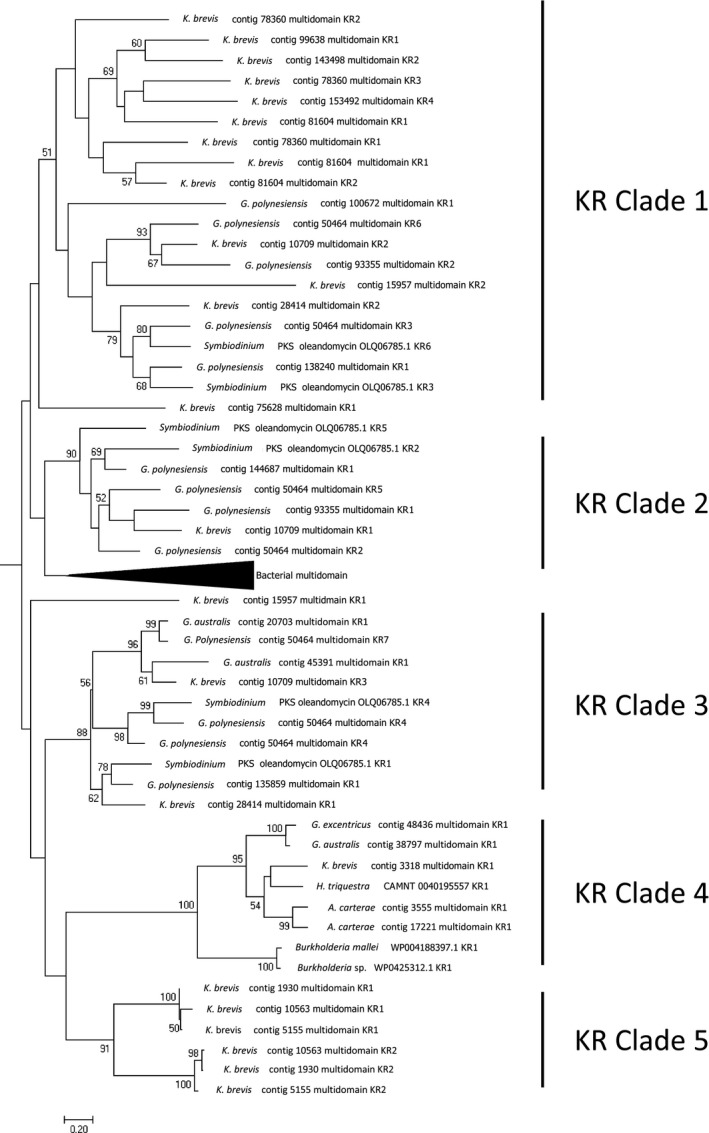
Details of clade membership of Karenia brevis KR domains summarized in Figure 5. KR domains from multidomain PKS sequences were found in subclades that correspond more closely to module architecture than to contig membership. Sub‐clade 1: KS‐KR‐ACP; Sub‐clade 2: KS‐DH‐KR; Sub‐clade 3 KS‐DH‐KR[ER]KR‐ACP: Sub‐clade 4: burA‐like NRPS/PKS; Sub‐clade 5: trans‐AT modules from NRPS/PKS with architecture KS‐DH‐KR in sequences with highly amplified ACP domains at their c‐terminal ends. G. australis, Gambierdscus australis; G. excentricus, Gambierdiscus excenctricus; H. triquestra, Heterocapse triquestra; A. carterae, Amphidinium carterae.
Survey of other PKS domains in Karenia brevis
Acyl transferase
Using HMMER, CD search and blast analysis we found 34 unique AT domain containing contigs (E value cut‐off ≤1E‐10). Of these, 9 occurred in the cis‐AT PKSs described above, while 25 encode single AT domains (Table S2). Phylogenetic analysis separates the cis‐AT domains from the stand‐alone ATs (Fig. S1 in the Supporting Information). Twenty single‐domain AT sequences have top blast hits of malonyl:acyl carrier transacylase. These sequences cluster separately from two sequences that are FabD domains of Type II FASs.
Dehydratase
By HMMER, CD search and blast analysis we found 24 unique DH domains, characterized by the presence of a “hotdog fold,” which consists of a seven‐stranded antiparallel beta‐sheet “bun” that wraps around a five‐turn alpha‐helical “sausage,” originally identified in E. coli FabA fatty acyl dehydratase (DH). The “hotdog fold” protein superfamily includes 17 subfamilies of proteins of diverse function including TEs and DHs (Dillon and Bateman 2004). Fourteen of the DH domains found are located within multidomain PKS or PKS/NRPS sequences (Table 3) with homology to either the “hotdog fold” superfamily or specifically to the PKS DH subfamily (Table S2). Another 9 contigs contain individual, partial “hotdog fold” domains (Table S2). Of these, one contains both spliced leader and polyA tail, indicating that it is not a partial sequence, but represents a full transcript. It is uncertain if these individual “hotdog fold” containing contigs function as standalone PKS DH since their “hotdog fold” domains are truncated. These sequences differ from the single Type II fatty acid beta‐hydroxyacyl‐ACP DH present in the assembly (Contig 44484), which has specific homology to the FabZ subfamily of “hotdog fold” superfamily.
Phosphopantetheinine attachment domains (PP)
PP attachment domains are conserved regions that are hallmarks of both ACPs in PKSs and PCP in NRPSs. HMMER analysis identified 123 contigs containing PP domains (E value cut‐off ≤1E‐3). Twenty of these are among the multidomain sequences described above (two are short contigs lacking PP domains). The majority of standalone PP domain contigs were <1,000 aa in length and contained 1 or 2 PP domains, with or without tpr repeat domains in the N‐terminal end. Several of these sequences possessed the dinoflagellate spliced leader, indicating their dinoflagellate origin.
Thioesterase domains
Using HMMER, and CD searches we found 49 TE domain containing contigs (E value cut‐off ≤1E‐3). Nine were among the multidomain sequences described above. Among the 40 standalone TE domain contigs (Table S2), several included a dinoflagellate spliced leader and in some cases polyA tails, indicating full length transcripts. Discrete TE domains (termed TEII) are frequently associated with multidomain PKS and NRPSs and play roles in editing and efficiency as well as chain release. Both integrated and discrete TE domains identified are members of the alpha/beta hydrolase fold class.
Unusual domains or motifs identified in K. brevis multidomain PKS and NRPK/PKS
Left‐handed parallel beta‐helix (LbH)
Regions with high similarity to LbH domains were found at or near the C‐terminal end of three multidomain PKSs: Contig 78360, Contig 15957, and Contig 54805 (Table 2). In contig 54805 three separate, nearly identical LbH motifs are found between pairs of ACP domains. Left handed beta helices are generated by 30 imperfect repeats of a hexapeptide motif (X‐[STAV]‐X‐[LIV]‐[GAED]‐X), where three contiguous repeats specify one turn of the β‐helix. Proteins containing hexapeptide repeats are often enzymes showing acyltransferase activity.
Thiamine pyrophosphate binding domain
A region with strong similarity to IlvB or other thiamine pyrophosphate binding domain (TPP) containing protein, was found at the N‐terminal end of Contig 34829, a hyrid NRPS/PKS (Table 2). IlvB is a transketolase involved in isoleucine synthesis that converts pyruvate to acetoacetate. Although unusual, TPP‐dependent transketolases have previously been reported to be the source of 2‐carbon donors from glycolytic pathway intermediates to NRPS assemblies (Peng et al. 2012).
Methyl transferase
Three similar contigs with trans‐AT architecture, Contig 1930, Contig 10563, and Contig 5155, possess a region at the C‐terminal end with homology to O‐methyltransferases. This family of MT utilizes S‐adenosyl methionine and have been shown in PKSs to incorporate methyl branches (Piel 2010). The location of the potential MT domain in these sequences, C‐terminal to a TE domain is unusual, as in Type I PKSs they usually occur in the AT‐KR, DH‐ER, or DH‐KR linker regions, or in a trans‐AT PKS between ACP domains (Ansari et al. 2008). The MT‐like regions in these contigs appear to be (identically) truncated and thus may not have activity.
Conclusions
Dinoflagellates produce an enormous diversity of polyketide compounds, many with adverse effects on human and marine animal health, yet their biosynthetic machinery has been little explored in large part because the considerable size of dinoflagellate genomes has hindered genomic sequence analysis. Over the past decade, transcriptome sequencing of a variety of dinoflagellates has revealed unique, single‐domain Type I PKSs likely to be involved in polyketide biosynthesis, while more typical multidomain PKSs have been consistently absent. In Karenia brevis, we (Monroe and Van Dolah 2008, Sanger sequencing) and others (Ryan et al. 2014, RNAseq, 50M reads/library) reported only single domain PKS transcripts. However, with RNAseq‐based access to deeper transcriptome sampling, as well as sequencing of the smallest dinoflagellate genome, Symbiodinium, there is increasing evidence for the presence of multidomain PKSs in dinoflagellates, in addition to the previously identified single domain PKS proteins. In the current study, we therefore pooled 20 RNAseq libraries to yield a combined library of 595M reads, in order to obtain sufficient coverage to assemble long, multidomain PKS transcripts if present. Using this approach, in addition to 121 single domain KS contigs, we found 22 contigs containing multiple KS domains, two NRPS sequences, and five hybrid NRPS/PKSs. Trans‐AT PKS architectures were prevalent, as recently reported in PKSs from other eukaryotic microalgae (Shelest et al. 2015). The longest PKS found in this study consisted of three modules, insufficient by itself to synthesize any of polyethers known to be produced by K. brevis. This suggests that multiple PKSs likely work together in the biosynthetic process. How, and if, the single domain KS sequences interact with multidomain PKSs is currently unknown, and is a question of great interest. Investigating the intracellular localization of the newly identified PKSs will be important for establishing potential interactions between single and multidomain sequences. The only dinoflagellate KS protein for which cellular localization has been investigated to date is K. brevis KB2006, a Clade 1 single domain KS, which was found in the chloroplast (Monroe et al. 2013). This study, in concert with recent evidence in Gambierdiscus spp. and Symbiodinium suggests that dinoflagellates utilize both single domain and multidomain PKS and PKS/NRPS proteins in their toxin biosynthetic machinery.
We thank Kelly Fridey Sides for generation of the heat stressed transcriptome and translatome libraries included in this analysis, with funding from the NOAA Marine Biotoxins Program. Support for this analysis was provided by the University of Technology Sydney Distinguished Visiting Scholar program to FMVD.
Supporting information
Figure S1. Phylogenetic analysis separates the cis‐AT domains from the stand‐alone ATs. Phylogeny was inferred using Mafft alignment and PHyML.
Table S1. Description of sequences from Karenia brevis encoding essential enzymes for various metabolic pathways (128 out of 133 enzymes are present). Enzymes colored in red were not present in the transcriptome.
Table S2. Description of all single‐domain contigs encoding KS, KR, DH, AT, ACP, and TE domains in the Karenia brevis assembly, and multidomain PKS and NRPS/PKS contigs containing incorporated modules. Contig sequences may be obtained from the NCBI Transcriptome Sequence Archive Accession number GFLM01000000.
References
- Ansari, M. Z. , Sharma, J. , Gokhale, R. S. & Mohanty, D. 2008. In silico analysis of methyltransferase domains in secondary metabolites. BMC Genom. 9:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff, T. R. , Williams, E. , Jagus, R. & Place, A. R. 2014. A non‐cryptic non‐canonical multi‐module NRPS/PKS found in dinoflagellates In MacKenzie A. L. [Ed.] Marine and Freshwater Harmful Algae 2014. Proceedings of the 16th International Conference on Harmful Algae. Cawthron Institute, Nelson, New Zealand: and the International Society for the Study of Harmful Algae (ISSHA), pp. 101–4. [Google Scholar]
- Baden, D. G. , Bourdelais, A. J. , Jacocks, H. , Michelliza, S. & Naar, J. 2005. Natural and derivative brevetoxins: historical background, multiplicity, and effects. Environ. Health Perspect. 113:621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, T. , Aranda, M. , Sunagawa, S. , Yum, L. K. , DeSalvo, M. K. , Lindquist, E. , Coffroth, M.A. , Voolstra, C.R. & Medina, M. 2012. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef‐building corals. PLoS ONE 7:e35269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedessee, G. , Hisata, K. , Roy, M. C. , Satoh, N. & Shoguchi, E. 2015. Multifunctional polyketide synthase genes identified by genomic survey of the synbiotic dinoflagellate, Symbiodinium minutum . BMC Genom. 16:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdelais, A. K. , Campbell, S. , Jacocks, H. , Naar, J. , Wright, J. L. C. , Carsi, J. & Baden, D. G. 2004. Brevenal is a natural inhibitor of brevetoxin action in sodium channel receptor binding assays. Cell. Mol. Neurobiol. 24:553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H. N. & Shimizu, Y. 1987. Biosynthesis of brevetoxins. Evidence for the mixed origin of the backbone carbon chain and the possible involvement of dicarboxylic acids. J. Am. Chem. Soc. 109:2184–5. [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J. M. , Terol, J. , Talón, M. & Robles, M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–6. [DOI] [PubMed] [Google Scholar]
- Davies, C. , Heath, R. J. , White, S. W. & Rock, C. O. 2000. The 1.8 Å crystal structure and active‐site architecture of β‐ketoacyl‐acyl carrier protein synthase III (FabH) from Escherichia coli . Structure 8:185–95. [DOI] [PubMed] [Google Scholar]
- Dillon, S. C. & Bateman, A. 2004. The Hotdog Fold protein superfamily includes 17 subfamilies of proteins of diverse function. BMC Bioinformatics 5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichholz, K. , Beszteri, B. & John, U. 2012. Putative monofunctional type I polyketide synthase units: a dinoflagellate‐specific feature? PLoS ONE 7:e48624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Clements, J. & Eddy, S. R. 2011a. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Clements, J. & Eddy, S. R. 2011b. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(suppl. 2):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, L. E. , Kirkpatrick, B. , Backer, L. C. , Walsh, C. , Nierenberg, K. , Clark, J. , Reich, A. et al. 2011. Review of Florida red tide and human health effects. Harmful Algae 10:224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, J. , Ishida, K. & Hertweck, C. 2012. Genomics‐driven discovery of a burkholderic acid, a non‐canonical cryptic polyketide from the human pathogenic Burkhorderia species. Angewandte Chem. Int. Ed. 51:11611–5. [DOI] [PubMed] [Google Scholar]
- Fridey, K. 2015. Bioinformatics approach to determining transcriptional and translational responses to heat stress in the Florida red tide dinoflagellate Karenia brevis. Thesis, College of Charleston, Charleston, SC, USA, 124 pp. [Google Scholar]
- Guillard, R. R. L. 1973. Divison rates In Stein J. [Ed.] Handbook of Phycological Methods ‐ Culture Methods and Growth Measurements. Cambridge University Press, Cambridge, UK, pp. 289–311. [Google Scholar]
- Guindon, S. , Dufayard, J. F. , Lefort, V. , Anisimova, M. , Hordijk, W. & Gascuel, O. 2010. New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–21. [DOI] [PubMed] [Google Scholar]
- Hardison, D. R. , Sunda, W. G. , Litaker, W. R. , Shea, D. & Tester, P. A. 2012. Nitrogen limitation increases brevetoxins in Karenia brevis (Dinophyceae): implications for bloom toxicity. J. Phycol. 48:844–58. [DOI] [PubMed] [Google Scholar]
- Hoagland, P. , Jin, D. , Polansky, L. Y. , Kirkpatrick, B. , Kirkpatrick, G. , Fleming, L. E. , Reich, A. , Watkins, S. M. , Ullmann, S. G. & Backer, L. C. 2009. The costs of respiratory illnesses arising from Florida Gulf Coast Karenia brevis blooms. Environ. Health Perspect. 117:1239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeckisch, N. , Yang, I. , Wohlrab, S. , Glockner, G. , Kroymann, J. , Vogel, H. , Cembella, A. & John, U. 2011. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii . PLoS ONE 6:e28012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke‐Kodama, H. , Sandmann, A. , Müller, R. & Dittmann, E. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22:2027–39. [DOI] [PubMed] [Google Scholar]
- John, U. , Beszteri, B. , Derelle, E. , Van de Peer, Y. , Readd, B. , Moreau, H. & Cembella, A. 2008. Novel insights into evolution of protistan polyketide synthases through phylogenomic analysis. Protist 159:21–30. [DOI] [PubMed] [Google Scholar]
- Kamykowski, D. , Milligan, E. J. & Reed, R. E. 1998. Biochemical relationships with the orientation of the autotrophic dinoflagellate Gymnodinium breve under nutrient replete conditions. Mar. Ecol. Prog. Ser. 167:105–17. [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K. I. & Miyata, T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellman, R. , Stüken, A. , Orr, R. J. S. , Svendsen, H. M. & Jakobsen, K. S. 2010. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 8:1011–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla, C. , Gokhale, R. S. , Jacobsen, J. R. & Cane, D. E. 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68:219–53. [DOI] [PubMed] [Google Scholar]
- Kimura, K. , Okuda, S. , Nakayama, K. , Shikata, T. , Takahashi, F. , Yamaguchi, H. , Skamoto, S. , Yamaguchi, M. & Tomaru, Y. 2015. RNA sequencing reveals numerous polyketide synthase genes in the harmful dinoflagellate, Karenia mikimotoi . PLoS ONE 10:e0142731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, G. S. , John, U. , Figueroa, R. I. , Rhodes, L. L. , Harwood, D. T. , Groth, M. , Bolch, C. J. S. & Murray, S. A. 2015. Polyketide synthesis genes associated with toxin production in two species of Gambierdiscus (Dinophyceae). BMC Genom. 16:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, G. S. , John, U. , Smith, K. , Fraga, S. , Rhodes, L. & Murray, S. A. 2017. Role of modular polyketide synthases in the production of polyether ladder compounds in the ciguatoxin‐producing Gambierduscus polynesiensis and G. excentricus (Dinophyceae). J. Euk. Microbiol. 64:691–706. [DOI] [PubMed] [Google Scholar]
- Kohli, G. S. , John, U. , Van Dolah, F. M. & Murray, S. A. 2016. Evolutionary distinctiveness of fatty acid and polyketide synthases in eukaryotes. ISME J. 10:1877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg, J. H. 2002. The effect of harmful algal blooms on aquatic organisms. Rev. Fisheries Sci. 10:113–390. [Google Scholar]
- Lee, M. S. , Qin, G. , Nakanishi, K. & Zagorski, M. G. 1989. Biosynthetic studies of brevetoxins, potent neurotoxins produced by the dinoflagellate Gymnodinium breve . J. Am. Chem. Soc. 111:6234–41. [Google Scholar]
- Lee, M. S. , Repeta, D. J. & Nakanishi, K. 1986. Biosynthetic origins and assignments of 13C NMR peaks of brevetoxin B. J. Am. Chem. Soc. 108:7855–6. [DOI] [PubMed] [Google Scholar]
- Lidie, K. L. , Ryan, J. C. , Barbier, M. & Van Dolah, F. M. 2005. Gene expression in the Florida red tide Dinoflagellate Karenia brevis: analysis of an expressed sequence tag (EST) library and development of a DNA microarray. Mar. Biotechnol. 7:481–93. [DOI] [PubMed] [Google Scholar]
- Lopez‐Legentil, S. , Song, B. , DeTure, M. & Baden, D. G. 2010. Characterization and localization of a non‐ribsomal peptide synthase and polyketide synthase gene from the toxic dinoflagellate Karenia brevis . Mar. Biotechnol. 12:32–41. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Derbyshire, M. K. , Gonzales, N. R. , Lu, S. , Chitsaz, F. , Geer, L. Y. , Geer, R. C. et al. 2014. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J. M. , Rödelsperger, C. , Eicholz, K. , Tillman, U. , Cembella, A. , McGaughran, A. & John, U. 2015. Transcriptomic characterisation and genomic glimpse into the toxigenic dinoflagellate Azadinium spinosum, with emphasis on polyketide synthase genes. BMC Genom. 16:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe, E. A. & Van Dolah, F. M. 2008. The toxic dinoflagellate Karenia brevis encodes novel type I‐like polyketide synthases containing discrete catalytic domains. Protist 159:471–82. [DOI] [PubMed] [Google Scholar]
- Monroe, E. A. , Johnson, J. G. , Wang, Z. , Pierce, R. K. & Van Dolah, F. M. 2010. Characterization and expression of nuclear encoded, chloroplast localized polyketide synthases in the dinoflagellate Karenia brevis. J. Phycol. 46:541–552. [Google Scholar]
- Morgan, K. L. , Larkin, S. L. & Adams, C. M. 2009. Firm‐level economic effects of HABS: a tool for business loss assessment. Harmful Algae 8:212–8. [Google Scholar]
- Patron, N. , Waller, R. & Keeling, P. 2006. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 357:1373–1382. [DOI] [PubMed] [Google Scholar]
- Parra, G. , Bradham, K. & Korf, I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 3:1061–7. [DOI] [PubMed] [Google Scholar]
- Pawlowiez, R. , Morey, J. S. , Darius, H. T. , Chinain, M. & Van Dolah, F. M. 2014. Transcriptome sequencing reveals single domain Type I‐like polyketide synthases in the toxic dinoflagellate Gambierdiscus polynesiensis . Harmful Algae 36:29–37. [Google Scholar]
- Peng, C. , Pu, J.‐Y. , Song, L.‐Q. , Jian, X.‐H. , Tang, M.‐C. & Tang, G.‐L. 2012. Hijacking a hydroxyethyl unit from a central metabolic ketose into a nonribosomal peptide assembly line. Proc. Natl Acad. Sci. USA 109:8540–8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, J. 2010. Biosynthesis of polyketides by trans‐AT polyketide synthases. Nat. Prod. Rep. 27:996–1047. [DOI] [PubMed] [Google Scholar]
- Prasad, A. V. K. & Shimizu, Y. J. 1989. The structure of hemibrevetoxinB: a new type of toxin in the Gulf of Mexico red tide organism. Am. Chem. Soc. 111:6476–7. [Google Scholar]
- Punta, M. , Coggill, P. C. , Eberhardt, R. Y. , Mistry, J. , Tate, J. , Boursnell, C. et al. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, D. E. , Pepper, A. E. & Campbell, L. 2014. De novo assembly and characterization of the transcriptome of the toxic dinoflagellate Karenia brevis . BMC Genom. 15:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo, T. , Upadhyay, R. J. , Nagasaki, K. & Bhattacharya, D. 2012. Dozens of toxin‐related genes are expressed in a nontoxic strain of the dinoflagellate Heterocapsa circularisquama . Mol. Biol. Evol. 29:1503–6. [DOI] [PubMed] [Google Scholar]
- Satake, M. , Campbell, A. , Van Wagoner, R. M. , Bourdelais, A. J. , Jacocks, H. , Baden, D. G. & Wright, J. L. C. 2009. Brevisin: an aberrant polycyclic ether structure from the dinoflagellate Karenia brevis and its implications for polyether assembly. J. Org. Chem. 74:989–94. [DOI] [PubMed] [Google Scholar]
- Shelest, E. , Heimeri, N. , Fichtner, M. & Sasso, S. 2015. Multimodular type I polyketide synthases in algae evolve by module duplications and displacement of AT domains in trans. BMC Genom. 16:1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, R. V. , Gibbs, P. D. L. , Palacios, A. , Abiy, L. , Dickey, R. , Lopez, J. V. & Rein, K. S. 2003. Polyketide synthase genes from marine dinoflagellates. Mar. Biotechnol. 5:1–12. [DOI] [PubMed] [Google Scholar]
- Snyder, R. V. , Guerrero, M. A. , Sinigalliano, C. D. , Winshell, J. , Perez, R. , Lopez, J. V. & Rein, K. S. 2005. Localization of polyketide synthase encoding genes to the toxic dinoflagellate Karenia brevis . Phytochemistry 66:1767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. 2006. RAxML‐VI‐HPC: maximum likelihood‐based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–90. [DOI] [PubMed] [Google Scholar]
- Staunton, J. & Weissman, K. J. 2001. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18:380–416. [DOI] [PubMed] [Google Scholar]
- Steidinger, K. A. 2009. Historical perspective on Karenia brevis red tide research in the Gulf of Mexico. Harmful Algae 8:549–61. [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. & Kumar, S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Bio. Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D. , Higgins, D. G. & Gibson, T. J. 1994. CLUSTAL W: improving the sensitivity of 642 progressive multiple sequence alignment through sequence weighting, position‐specific gap 643 penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truxall, T. , Bourdelais, A. J. , Jacocks, H. , Abraham, W. M. & Baden, D. G. 2010. Characterization of tamulamides A and B: polyethers isolated from the marine dinoflagellate Karenia brevis . J. Nat. Prod. 73:536–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiner, M. , Flewelling, F. , Fire, S. , Bowen‐Stevens, S. , Gaydos, J. , Johnson, C. , Landsberg, J. et al. 2012. Comparative analysis of three brevetoxin‐associated bottlenose dolphin (Tursiops truncatus). PLoS ONE 7:e42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wagoner, R. M. , Satake, M. , Bourdelais, A. J. , Baden, D. M. & Wright, J. L. C. 2010. Absolute configuration of brevisamide and brevisin: confirmation of a universal biosynthetic process for Karenia brevis polyethers. J. Nat. Prod. 73:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dolah, F. M. , Zippay, M. , Pezzolesi, L. , Rein, K. , Johnson, J. G. , Wang, Z. & Pistocchi, R. 2013. Subcellular localization of polyketide synthases and fatty acid synthase activity in dinoflagellates. J. Phycol. 49:1118–1127. [DOI] [PubMed] [Google Scholar]
- Van Wagoner, R. M. , Satake, M. & Wright, J. L. 2014. Polyketide biosynthesis in dinoflagellates: what makes it different? Nat. Prod. Rep. 31:1101–37. [DOI] [PubMed] [Google Scholar]
- Wright, J. L. C. , Hu, T. , McLachlan, J. L. , Needhm, J. & Walter, J. A. 1996. Biosynthesis of DTX‐4: confirmation of a polyketide pathway, proof of a Baeyer‐Villiger oxidation step, and evidence for an unusual carbon deletion process. J. Am. Chem. Soc. 118:8757–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic analysis separates the cis‐AT domains from the stand‐alone ATs. Phylogeny was inferred using Mafft alignment and PHyML.
Table S1. Description of sequences from Karenia brevis encoding essential enzymes for various metabolic pathways (128 out of 133 enzymes are present). Enzymes colored in red were not present in the transcriptome.
Table S2. Description of all single‐domain contigs encoding KS, KR, DH, AT, ACP, and TE domains in the Karenia brevis assembly, and multidomain PKS and NRPS/PKS contigs containing incorporated modules. Contig sequences may be obtained from the NCBI Transcriptome Sequence Archive Accession number GFLM01000000.


