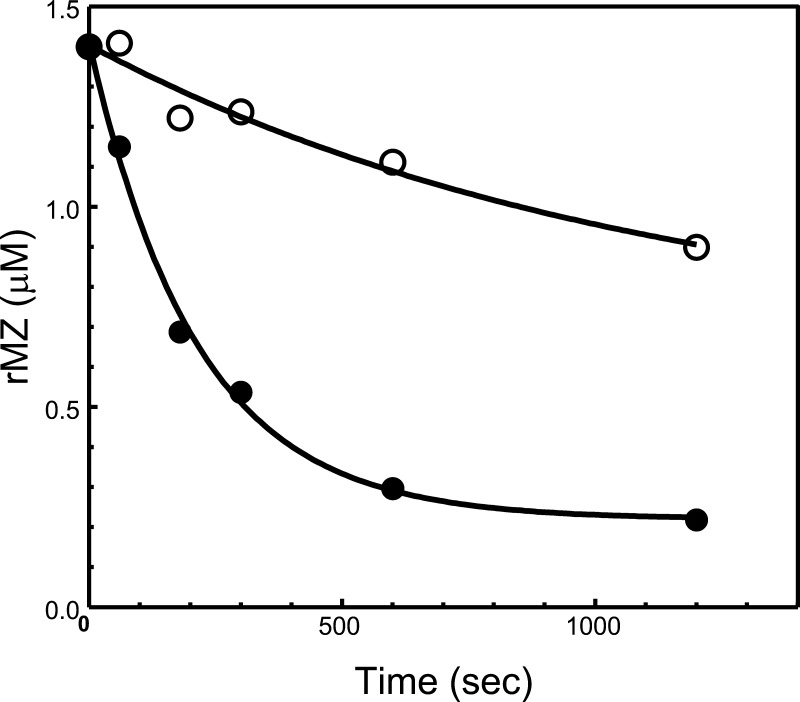
Figure 6.
Analysis of rMZ-II consumption by prothrombinase assembled with rhfVa molecules. The gel shown in Figure 5 (A) was scanned and hPro consumption was recorded as described in the Experimental Section. Following scanning densitometry, the data representing recombinant mutant hPro consumption as a function of time (s) were plotted using nonlinear regression analysis according to the equation representing a first-order exponential decay using the software Prizm (GraphPad, San Diego, CA), as described in the Experimental Section. Prothrombinase was assembled with rhfVaWT (filled circles) or rhfVaNR→DE (open circles). The apparent first-order rate constant, k (s–1) was obtained directly from the fitted data, and the resulting numbers representing recombinant mutant hPro consumption are reported in Table 2.