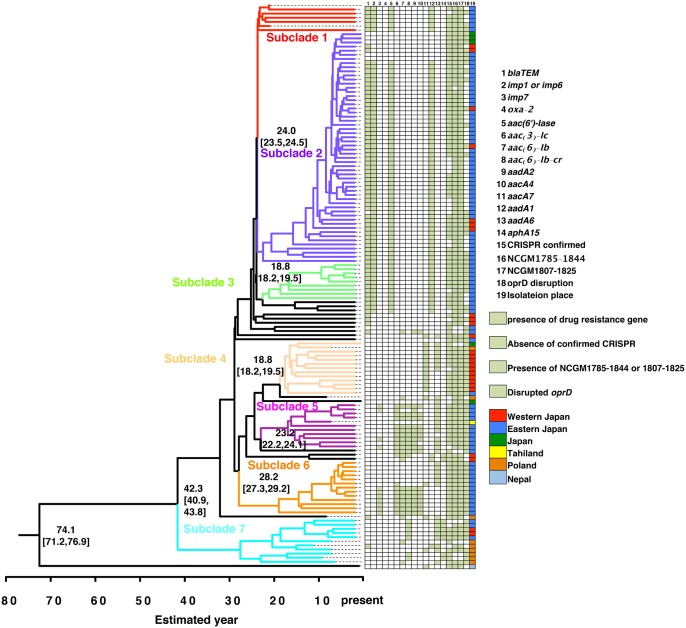
Fig. 2.
—Phylogenic tree of the 136 ST235 Pseudomonas aeruginosa strains. The phylogeny of ST235 MDR P. aeruginosa strains was evaluated using the BEAST program (Drummond and Rambaut 2007). The seven subclades are shown in color. The estimated ages of the branches are shown as median values with 95% highest posterior density (HPD). The proportion of isolates from each location carrying conserved antibiotic resistance genes (>50% per subclade) and their oprD disruption status are also indicated. The posterior probability value for each main branch was >0.99. More detailed results are shown in supplementary figure 2 and supplementary data 1, Supplementary Material online. The posterior probability of each subclade designated was > 0.95.