Abstract
Although more and more entangled participants of translation process were realized, how they cooperate and co-determine the final translation efficiency still lacks details. Here, we reasoned that the basic translation components, tRNAs and amino acids should be consistent to maximize the efficiency and minimize the cost. We firstly revealed that 310 out of 410 investigated genomes of three domains had significant co-adaptions between the tRNA gene copy numbers and amino acid compositions, indicating that maximum efficiency constitutes ubiquitous selection pressure on protein translation. Furthermore, fast-growing and larger bacteria are found to have significantly better co-adaption and confirmed the effect of this pressure. Within organism, highly expressed proteins and those connected to acute responses have higher co-adaption intensity. Thus, the better co-adaption probably speeds up the growing of cells through accelerating the translation of special proteins. Experimentally, manipulating the tRNA gene copy number to optimize co-adaption between enhanced green fluorescent protein (EGFP) and tRNA gene set of Escherichia coli indeed lifted the translation rate (speed). Finally, as a newly confirmed translation rate regulating mechanism, the co-adaption reflecting translation rate not only deepens our understanding on translation process but also provides an easy and practicable method to improve protein translation rates and productivity.
Keywords: translation rate, co-adaption, amino acid usage, tRNA gene copy number, minimum cost
1. Introduction
Translation initiation, elongation and termination involve many factors, that balance translation rate (speed) and accuracy.1,2 The final translation efficiency for a given protein is restricted by the cost of its production and organization.3 Therefore, evolving a genome-wide translation regulation regime can efficiently determine the translation rates of various genes under different conditions.1 Conventional computations of translation elongation efficiency refer to codon usage4 and tRNA availability.5 The relationship between codon usage and tRNA abundance predicts translation efficiency with reasonable accuracy.1
Additional theories have been proposed with constantly emerging experimental technologies6,7 to cope with challenges to the simplified assumptions about translation described above. Thus, the effects of codon order,8–10 local tRNA availability,11–13 regulation of expression of the tRNA gene,14 the diverse demands of the transcriptomes,9,15 ribosomes,16 mRNA structures17–19 and folding energy20 were included in the translation efficiency models. Among these factors, tRNA availability repeatedly emphasized decides the supply of aminoacyl-tRNA,21 which influences the translocation of ribosomes on mRNA.22–25 Nutriment limitations, such as amino acid shortage, also have influences on the cellular supply of aminoacyl-tRNA.26 However, how the formation of aminoacyl-tRNA influences translation efficiency is still unclear.
In the translation process, tRNAs can be thought of as tools and the amino acids as the raw materials. Each species of tRNA corresponds to a particular amino acid, and each of the former is responsible for carrying one of the latter. We hypothesize that the levels of the tRNAs and the corresponding amino acids should be well matched to synthesize proteins more efficiently. Such a consistency would maximize efficiency and minimize resource/energy costs.
Here, we firstly examined the association between the tRNA gene copy numbers and amino acid compositions in various organisms. We sought to validate two points of reasoning: at the organismal scale, most organisms evolve co-adaption between tRNA gene copy number and amino acid composition; and at the second scale of individual proteins, the co-adaption intensity may vary among the proteins within an organism. Some proteins need to be expressed rapidly to maintain a high quantity or to satisfy the requirements of acute responses. We speculated that such proteins would have higher co-adaption to increase their translation efficiency. Computational analyses were employed to elucidate co-adaption between tRNA gene copy numbers and amino acid usage for proteins/proteomes in three domains of life, indicating the effects of maximum efficiency and the minimum cost principle. Then, we further correlated the co-adaption with proteins’ translation rates, which were validated by growth rates of bacteria and production rates of target proteins. The target proteins’ translation rates were observed to be lifted through changing the proportions of gene copy numbers of tRNAs, providing clues for applied biology.
2. Materials and methods
2.1. E. coli strain and methods
DNA amplification and expression were performed in E. coli Top10 cells (F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 Δ lacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG). All bacterial media and methods used in this study were as described in Current Protocols in Molecular Biology.27
2.2. Production of synthetic genes
Oligonucleotides were synthesized using PCR amplification. The fragments were recombined to generate the target coding sequences, which were inserted behind the arabinose promoter. Positive clones were screened by resistance screening and confirmed by PCR and sequencing.
2.3. Detection of target polypeptides
Cells were grown overnight at 30°C in Luria–Bertani (LB) culture medium, and were inoculated in LB culture medium with ampicillin at OD600 = 2. After hours of constant shaking (OD600 = 0.6), L-arabinose (0.05%) was added to the culture medium to induce heterologous expression. Samples were collected at different time points and put on ice. When all samples were prepared, aliquots of the cells were observed through confocal microscopy (Leica TCS SP8, Germany), and the rest were collected by centrifugation (4000×g, 20 min). The cells were then washed three times with cold PB (4000×g, 10 min), and cell lysis buffer [phenylmethylsulfonyl fluoride (PMSF) 0.1 mM, PB 10 mM, lysozyme 1 mg/ml] was added to lyse the cells for 15 min before sonication (3 min). After ultrasonic breakage, the samples were centrifuged, and the supernatants were collected. EGFP in the supernatants was measured using fluorospectro-photometer (Hitachi F7000, EX WL: 460.0 nm) and was quantified using a Bicinchoninic Acid (BCA) Protein Assay Kit (CWBIO) before adding loading buffer and boiling for five minutes. Thirty micrograms of the protein extract samples were loaded on 12% stain-free SDS-polyacrylamide gel (Bio-Rad), subjected to electrophoresis, and transferred to 0.2 µm polyvinylidene difluoride membranes (Millipore). After blocking with 5% non-fat milk in Tris-buffered saline buffer with Tween 20 (TBS/T) for 4 h at room temperature, the membrane was incubated with the appropriate primary antibodies (Abmart GFP-tag mAb, 1:1000) for 18 h at 4 °C. Next, horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody (1:5000, ZSGB-BIO) was applied for 2 h at room temperature. Finally, the signals were visualized using an Enhanced Chemiluminescence (ECL) kit (Roche).
2.4. TAAI
TAAI is short for the ‘tRNA gene copy number and amino acid usage accordance index’, and it measures the co-adaption of amino acid usage and tRNA gene copy number. For a given protein sequence, the frequencies of the 20 types of amino acids are unique. For a specific organism, the average frequencies of amino acid usage also differ among organisms.
| (1) |
| (2) |
| (3) |
where Xi is the frequency of the gene copy of tRNA i in one organism’s genome in Equation (1), and let Yi be the frequency of amino acid i of in a specific protein in the Equation (2). The value NtRNAi is the corresponding gene copy number of tRNAi decoding all codons for the ith amino acid. The value Naai is the corresponding counts of the ith amino acid in special protein or genome. TAAI in Equation (3) is the Spearman correlation index of X and Y. In addition to the TAAI of one protein, we also calculated the general TAAI of an organism to denote its overall co-adaption. The overall value equals the average of all proteins’ TAAIs for that organism.
2.5. Bioinformatics data source
This work requires tRNA gene annotation information that is as accurate as possible; therefore, we compared annotation information from three databases. We chose three widely used databases: GenBank,28 a comprehensive bioinformatics database; the Genomic tRNA Database,12 which uses tRNAscan-SE;29 and tRNAdb,30 which contains more than 12,000 tRNA genes. A total of 410 genomes have the same tRNA gene annotation information in all three databases. If the tRNA gene annotations for one organism were consistent in the three databases, we deemed it as reliable and then employed this organism in further analysis. Nucleotide and protein sequences were downloaded from GenBank,28 and protein abundance values were acquired from PaxDb.31 We further compared the co-adaption of specific gene categories. Further analysis employed housekeeping genes (expression-invariable ones) and tissue-specific genes (expressed only in one tissue) from Ma et al.32 The young and old genes of human were acquired from Yin et al.33 and Wei et al.34 The horizontal transfer gene lists for bacteria were downloaded from HGT-DB.35
We employed 53 organisms’ growth times (Supplementary Table S2). Here, we chose those archaea and bacteria with growth times from Rocha et al.36 Among 410 organisms filtered for reliable tRNA gene annotation, 376 archaea and bacteria were chosen to perform the analysis of correlation between TAAI and genome sizes. When comparing the TAAIs among proteins within a specific genome, we analyzed the protein abundances and TAAIs in six model organisms, which have integrated abundances in PaxDb.31
3. Results
3.1. At the genome scale the tRNA and corresponding amino acid co-adapt to maximize the efficiency
During translation, tRNAs transport amino acids to ribosomes, and co-operation between the two components has been reported in a few organisms.37 Previous researches demonstrated that the tRNA gene copy numbers were different among organisms/strains/species, and that the protein sequences varied greatly.38 To check whether the amino acid usage of proteins generally co-adapts with the corresponding organism’s tRNA gene copy number, we calculated and compared independent frequencies of the two in 410 genomes from three domains of life (17 archaea; 359 bacteria; 34 eukaryotes), using more accurate tRNA gene annotations in GenBank,28 the Genomic tRNA database12 and tRNAdb.30 First, the frequencies of 20 standard tRNA genes (Supplementary Table S1) were computed by counting cognate tRNA gene copies and being divided by the organism’s total tRNA gene counts. Second, frequencies of the 20 amino acids in the proteome (Supplementary Table S1) were computed by dividing the count of each amino acid by the sum of the twenty amino acid counts. After obtaining these two types of data for each organism, we performed linear fit and correlation analyses.
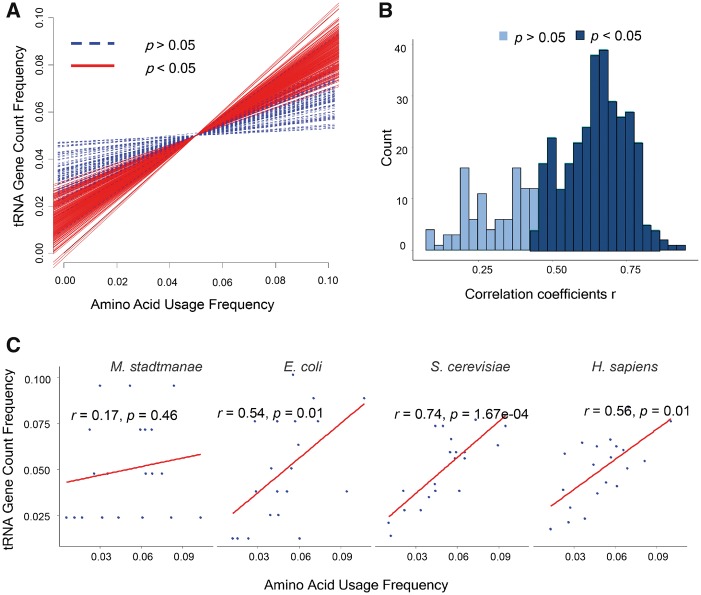
The linear fit results showed variable co-adaption relationships (Fig. 1A and Supplementary Table S1), illustrating that the two factors (tRNA gene copy number and the amino acid frequency) are not independent from each other. Indeed, although the slopes of the fitted lines differed, in all cases, the tRNA gene copy numbers showed positive correlations with corresponding amino acid usages. Spearman rank correlation coefficients (r) were calculated after least square fitting (Supplementary Table S1). Specifically, 99.27% had correlation coefficients greater than 0.1 (Fig. 1B), and 75.61% showed significant correlations (P < 0.05). Finally, a general linear relationship exists between tRNA gene copy and amino acid usage. For four representative organisms, the archaebacterium Methanosphaera stadtmanae (r = 0.17, P = 0.46), the bacterium Escherichia coli (r = 0.54, P = 0.01), and the eukaryotes Saccharomyces cerevisiae (r = 0.74, P = 1.67e−04) and Homo sapiens (r = 0.56, P = 0.01), the linear models presented different co-adaption intensities (Fig. 1C and Supplementary Table S1). Compared with the other three organisms, yeast had the best linear fit. However, for M. stadtmanae, having the most unbalanced constitution of tRNA genes, the observed points were not well fitted. In general, most organisms’ tRNA gene copy numbers and amino acid usages showed a positive linear relationship.
Figure 1.
Co-adaption between frequencies of tRNA gene copy numbers and amino acid usage. (A) Linear fitting results for 410 organisms. Dotted lines represent organisms with the P values of linear fitting are greater than 0.05; straight lines indicate that P values less than 0.05. (B) Corresponding Spearman rank correlation coefficients r for the linear fit of 410 genomes. (C) Linear fit of four model organisms.
We measured the co-adaption intensity using a tRNA gene copy and amino acid usage accordance index (TAAI), which equals to the r value of the Spearman rank correlation. During protein production, tRNA genes will be transcribed to tRNAs, and then loaded with amino acids for protein translation. Resource allocation would be the most efficient if the supply of tRNAs just meets the required amount of amino acids. Based on the results, our species/strain/organism scale reasoning was confirmed. In other words, most genomes had significant co-adaption between the tRNA gene copy number and the frequencies of amino acid usage and hence maximized their translation efficiency and minimized their energy/resource costs.
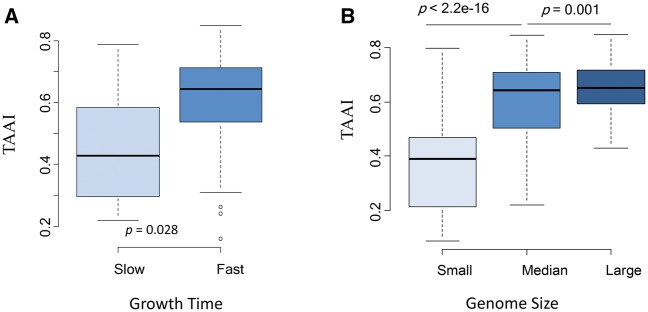
Different genomes (species/strain/organism) may face different translation selection pressure when translating different numbers of proteins in a given time. For example, large genomes have more proteins, and fast-growing bacteria need to synthesize more proteins simultaneously. In fact, large bacterial genomes are often associated with short generation times.36 According to the maximum efficiency/minimum cost principle, fast-growing/large bacteria should have higher TAAIs than slow-growing and/or small bacteria. To test this possibility, we compared the TAAIs of 53 bacteria (Supplementary Table S2) and grouped them by growth time.36 The fast, had growth times below the mean of the 53 ones; the slow, had growth times greater than the average. The two groups had similar variances and GC contents, while the slow group had significantly lower TAAIs than the fast group (Fig. 2A). Thus, co-adaption showed an effect on growth rate. This result is consistent with the idea that population growth rate is a fundamental ecological and evolutionary characteristic of living organisms.39 Similarly, larger bacteria have larger genomes and more proteins that need to be translated than bacteria with smaller genomes.39,40 Therefore, we also compared the TAAIs of prokaryotic organisms grouped by genome size (small, medium and large). These three groups had significantly divergent mean TAAIs of 0.37, 0.60 and 0.65 (the Student’s t test: P < 2.2e−16; Fig. 2B). That the relatively larger genomes have higher TAAIs supports the conclusion that bacteria under higher selective pressure have higher TAAIs, thus conforming to our first hypothesis based on the principle of efficiency described above. Bacteria with smaller genomes and slow growing speeds would suffer less pressure from protein translations and hence have lower TAAIs.
Figure 2.
Co-adaption at the genome scale. (A) TAAIs of bacteria divided into two groups based on their growth rates. The dataset includes 53 bacteria with available information on growth rates: which were classed into two groups by the mean growth time. The fast group has higher TAAIs than the slow. The GC content of the two groups are similar (The students’ t test: P = 0.42), their genome sizes varied non-significantly (The fast generally has larger genome than the slow: P = 0.07). (B) Prokaryotic organisms’ TAAIs, associated with corresponding genome sizes. The prokaryotic organisms were divided into three groups, showing significantly different TAAIs. Here, 376 prokaryotic genomes were involved in the analysis of correlation between TAAIs and genome sizes. In addition, the GC content also affect the TAAIs along with the increase of genome sizes: GC small < GC median < GC large (P < 2.2e−16).
Some genomes have non-significant TAAIs and it would be beneficial to understand why. In prokaryotes, genome size and TAAI correlated well (r = 0.49, P < 2.2e−16) and almost all the 96 prokaryotes with bad TAAI/co-adaption do have genome sizes smaller than 2.5 Mb, whereas almost all genomes with good co-adaption are larger than 2.6 Mb. The weak TAAI of smaller genomes is obviously caused by their deficiency in request of translation efficiency: less selection pressure, which could be measured based on the genome size in these organisms. However, in eukaryotes, alternative splicing of messenger RNA results in an inconsistency between genome size and the number of proteins produced. Using the quotient of the number of proteins divided by genome size should be a more reliable reflection of eukaryotes’ actual translation demand (also selection pressure). Consequently, four eukaryotes, Bos taurus (cow), Felis catus (cat), Strongylocentrotus purpuratus (Sea urchin) and Plasmodium falciparum with non-significant TAAIs indeed have smaller quotient. Therefore, it is reasonable that the lower translation demand (also selective pressure) leads some genomes to have bad TAAIs.
3.2. Highly expressed proteins and those connected to acute response tend to have higher TAAI for fast production
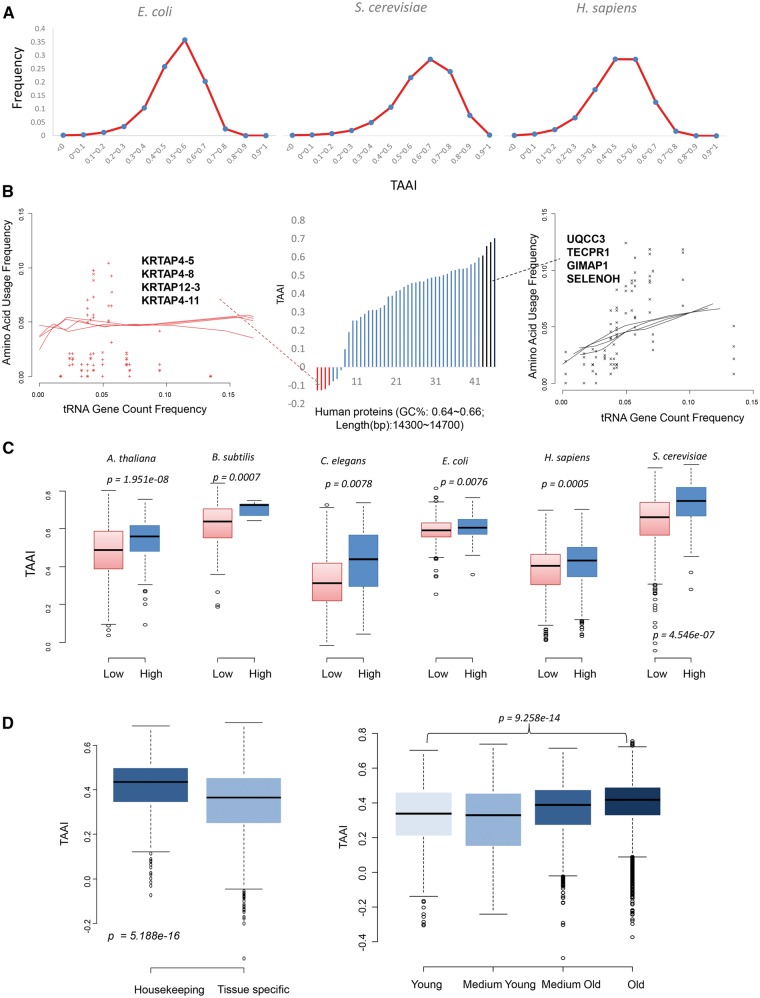
The aforementioned results validated the maximum efficiency principle at the genome (species/strain/organism) scale. Next, we asked whether there are co-adaption divergences within genomes and what such divergences may signify. The proteins within a genome also have different adaptions (variable TAAIs) for their different amino acid compositions (Fig. 3A). Taking H. sapiens as an example, a distinct difference was noted when comparing the co-adaption of the four proteins with the highest TAAIs and the ones of four proteins with the lowest TAAIs, which belong to a gene set with similar GC content and gene lengths (Fig. 3B). The amino acid frequencies of the top four were more consistent with the corresponding genomic tRNA gene copy frequencies. Within a given genome, this co-adaption divergence generally occurred among proteins.
Figure 3.
Intra-genome variation and translation factors. (A) TAAI distribution in three model organisms. (B) For human proteins having similar backgrounds (GC%: 0.64–0.66; lengths: 14,300–14,700 bp), they have variable intensities of co-adaption. Amino acid compositions of the top four proteins (Black ones) linearly correlated with the tRNA gene copy numbers, and Amino acid compositions of the bottom four (Red ones) do not change with the growth of the tRNA gene copy numbers, the gene names were listed in the subfigures. (C) Analysis of six model organisms’ abundance shows that highly expressed proteins generally have higher TAAIs than proteins with lower expression levels. Proteins in the high and the low groups have the similar GC content and lengths (The students’ t test: P > 0.05). Their GC contents are: A. thaliana 0.41–0.42; C. elegans 0.34–0.35; S. cerevisiae 0.39–0.41; E. coli 0.50–0.53; B. subtilis 0.50∼0.53; H. sapiens 0.5–0.53. We try to determine the most appropriate GC contents under which more proteins can be included into the analysis. (D) The housekeeping genes and old genes have higher TAAIs than those tissue specific and young genes. These results accord with the results by Ma et al. and Yin et al. that housekeeping and old genes suffered more translation selection.
Selective pressure within genomes is reflected by the direct results of translation efficiency: proteins’ abundances, even though the two values are not entirely equivalent. Co-adaption should reflect the supply of aminoacyl-tRNA, which ultimately affects the final protein synthesis. We compared six model organisms’ TAAIs and found that proteins with higher expression levels had clearly higher TAAIs than proteins with lower expression levels by Student's t test, even these proteins are with similar GC content and gene lengths (Fig. 3C). Furthermore, when using a linear fit, the TAAIs and the protein abundances showed significantly positive correlations (P < 1e−6; Supplementary Table S3). This result is consistent with the idea that tRNA level has direct effects on translation efficiency.41 Thus, as a reflection of the translation rate, protein abundances correlate with TAAIs to a certain extent, and their relationship seems to be a consequence of selection pressure to keep a suitable translation rate.
To further explore this finding, and considering that paralogous genes in the same family have similar molecular evolutionary stresses and changes,42 we compared the TAAIs in E. coli and yeast according to gene function groups: ribosome subunits, cell division,43 two-component system (including response regulators and sensors,44 mismatch repair45 and sugar metabolism.46 The average TAAIs and protein abundances for five groups of genes were calculated (Table 1). For the E. coli and yeast genomes, proteins from all these groups had average TAAIs higher than the genome average (Student’s t test: P = 3.54e−12). Three of the five groups correspond to acute responses (cell division, two-component system, and mismatch repair), and the other two groups are related to fast growth (ribosome subunits and sugar metabolism). Ribosome subunits are important participants in the translation process, and there is no doubt that ribosome subunits have the highest abundance, TAAI and codon usage bias index CAI.4 Sugar metabolism proteins, which includes proteins in ‘Amino sugar and nucleotide sugar metabolism’, also has higher TAAI and abundance than genome average. Although the abundances of proteins in the other three categories are lower than genome average, their TAAIs were higher. We further compared the protein abundance and TAAIs of experimentally determined upregulated yeast genes,47 and observed similar results (Supplementary Table S4). Thus, proteins involved in acute responses under more selective pressure, generally have good co-adaption relationships.
Table 1.
Analysis of the potentially rapidly expressed proteins according to their functions in E. coli and S. cerevisiae
|
E. coli |
S. cerevisiae |
|||||||
|---|---|---|---|---|---|---|---|---|
| Number | Abundance | TAAI | CAI | Number | Abundance | TAAI | CAI | |
| Whole genome | 3133 | 319.2 | 0.51 | 0.35 | 6087 | 163.8 | 0.62 | 0.18 |
| Ribosome subunits | 57 | 4724.28 | 0.59 | 0.63 | 178 | 1231.65 | 0.74 | 0.48 |
| Cell division | 19 | 131.84 | 0.57 | 0.35 | 20 | 15.09 | 0.67 | 0.16 |
| Two-component system | 58 | 80.7 | 0.57 | 0.29 | 216 | 203.86 | 0.65 | 0.19 |
| Mismatch repair | 20 | 57.21 | 0.53 | 0.34 | 19 | 32.51 | 0.72 | 0.16 |
| Sugar metabolism | 47 | 325.52 | 0.52 | 0.41 | 28 | 563.53 | 0.69 | 0.26 |
Proteins in bold face had lower corresponding values than genome’s average values.
According to Yin et al.33 and Ma et al.,32 old genes and housekeeping experience stronger translational selection. Thus, the co-adaption in these genes should be better. We employed the gene lists from this two papers and arrived at the expectant conclusion (Fig. 3D, Supplementary Table S5). Additionally, those horizontally transferred genes experiencing less translation selection also have bad co-adaptions (Supplementary Fig. S1). Therefore, our second reasoning was also validated: at the scale of individual proteins, co-adaption intensity may vary among the protein collective within a genome. Some proteins need to be expressed rapidly to maintain their quantity to be connected with an acute response.
3.3. Experimentally optimizing co-adaption indeed lift the protein expression
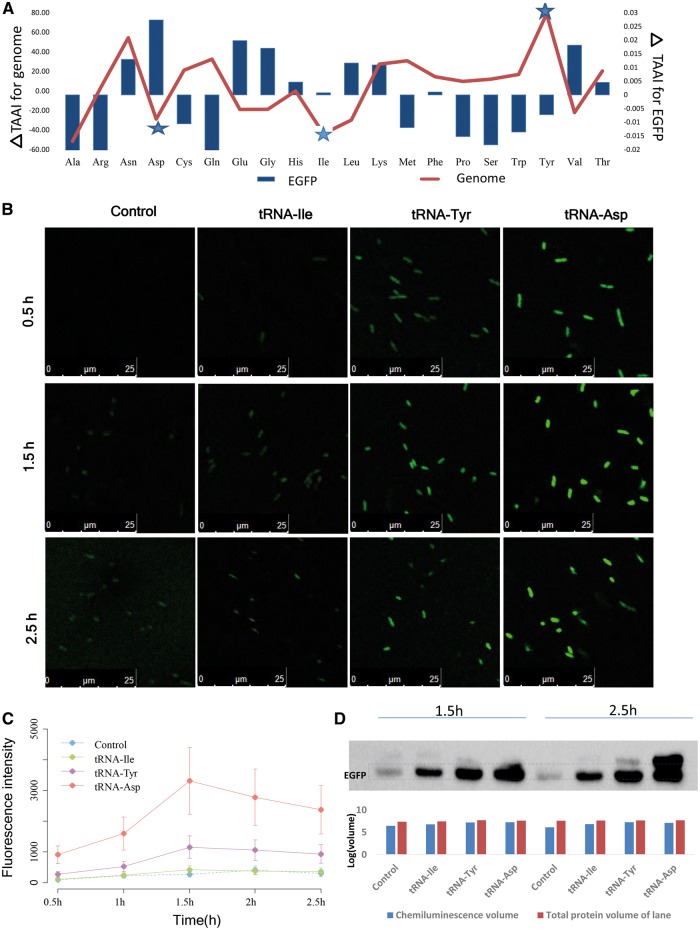
According to the principle and the results above, co-adaption was associated with the translation rate, prompting us to examine whether the proteins with high TAAIs indeed have high translation rates in vivo. To test this conjecture, one copy of a specific tRNA gene that might increase or decrease the TAAI was introduced to E. coli along with a gene encoding enhanced green fluorescent protein (EGFP) after which EGFP protein synthesis was analyzed.
Proteins with higher TAAIs might have higher translation rates, and thus higher production levels. EGFP is easy to express and detect, and constructs for tRNA overexpression have previously been designed and tested.48 Therefore, by combining the expression of EGFP with a tRNA gene, we can detect the effect of a specific tRNA on EGFP expression. Increasing the copy number of the corresponding tRNA gene may increase or decrease the TAAI for EGFP and the whole genome (Fig. 4A). EGFP had a TAAI of 0.45 when expressed with the original frequencies of E. coli tRNA genes. When one gene copy encoding either , or was introduced, the corresponding for EGFP were 0.03, 0.0008 or −0.007, respectively, and the cumulative for all proteins of the genome were −28.68, −43.77 or 78.68, respectively. The following sequences were constructed in plasmids: (control group), (tRNA-Asp), (tRNA-Ile) and (tRNA-Tyr), which were used to transfer E. coli Top10 cells (Fig. 4B). In E. coli, there is only one type of tRNA (isoacceptor tRNA) for Asp, Tyr and Ile, which means that codon bias has no observable effect on the increase in EGFP expression. We observed fluorescence intensity with confocal microscopy and found that the EGFP yield of the experimental group expressing was significantly higher than the others at the same time point (Fig. 4B and Supplementary Fig. S2A). Detailed fluorescence intensities of the four groups acquired with a fluorospectro photometer (Fig. 4C), showed that the EGFP production efficiency of the four groups from low to high was as follows: Control, tRNA-Ile, tRNA-Tyr and tRNA-Asp. The fluorescence intensities were consistent with western blotting results (Fig. 4D and Supplementary Fig. S2B). The experimental group tRNA-Asp produced ten times more EGFP than the Control. Considering the dynamic processes involved in EGFP abundance variation, the slope of abundance was much higher than that of the control, indicating that the translation rate of the former was higher.
Figure 4.
EGFP expression of original and optimized TAAIs in E. coli. (A) is the upregulated TAAI value resulting from adding a copy of one of the 20 standard tRNAs. The star marks tRNA-Asp, tRNA-Ile and tRNA-Tyr. (B) Confocal micrographs of the control and experimental groups present different fluorescence intensities in the EGFP channel; the corresponding merge figures of the bright-field and EGFP channel are shown in Supplementary Fig. S2. (C) Fluorescence intensities of four nascent sequences with EGFP at 513 nm from 0 to 2.5 h. All of these results showed that in the experimental group transformed with the Asp tRNA gene, there was a lift approximately ten-fold. (D) Western blot results for the nascent sequences. The following histograms show the normalized density of the corresponding lane, and the chemiluminescence intensity of the corresponding target band using stain-free technology (Bio-Rad). The corresponding electrophoretogram, shown in Supplementary Fig. S2B, reflects the loading volume of the total proteins.
To further rule out the possibility of codon usage influence, we compared the number and coding order of codons for the three tRNAs in EGFP (Supplementary Fig. S2C). The EGFP mRNA sequence has one type of Ile codon (ATC), two types of Tyr codons (TAT: 9%, TAC: 91%), and two Asp codons (GAT: 11%; GAC: 89%). All preferred codons are the corresponding codons for the E. coli cognate tRNAs. Therefore, there should be no significant variant effect of varying from the preferred codon. Then, we calculated the dispersion degree by analysis of variance, as the order of tRNA can influence its recycling during translation.14 We analyzed the variance of amino acid sites both locally and globally. The variance of the first eleven sites for Ile, Tyr and Asp are: 53, 61 and 46. The corresponding recycling effects should be weakened when increasing specific tRNA gene copy numbers. In fact, increasing the Ile tRNA gene copy number does not significantly increase EGFP production. Together, the results do not indicate an influence of tRNA recycling or preferred codon usage. This experiment confirmed that co-adaption has a clear effect on translation rate. Thus, optimizing co-adaption could significantly promote translation production of foreign proteins.
4. Discussion
4.1. This co-adaption relationship benefits translation rate differently from codon usage
Cells are believed to evolve to maximize efficiency and minimize resource and energy cost.49,50 We hypothesized that this principle would affect translation mechanisms, and we tested this conjecture based on the basic translation ‘tool’ tRNAs and the ‘raw material’ amino acids. To maximally utilize the resources, we reasoned that the quantities of the 20 tRNAs and amino acids in a species should be consistent based on this principle. For simple and convenient analysis, we used the tRNA gene copy number as the proxy for the former, and used the amino acid frequency as the latter. The genome has an average vector of amino acid frequency and each protein also has its vector form of amino acid frequency. Hence, there would be a general co-adaption value for each genome and a specific co-adaption value for each protein. Using correlation and abundance (functional group) analyses we validated our two conjectures, which are logical outcomes of the maximum efficiency and minimum cost principle. Based on the results and analyses, the genome’s TAAI could be regarded as a proxy for general translation efficiency or actual translation needs. Proteins’ TAAIs reflect the highest translation efficiency or translation need in extreme conditions.
Co-adaption is a global effect exerted on proteins and organisms. Each organism has a specific amino acid usage and a co-adapted tRNA gene copy number. This co-adaption maximizes the translation efficiency of the complete proteomes. The larger translation pressure the organism is exposed to, the higher average TAAI it has. In a genome, almost all the proteins have positive TAAI values (Fig. 3A). Hence, this co-adaption as a translation rate associated factor is applicable to all three domains of life and all proteins within an organism. However, facing multi rate-limiting factors, it is uncertain how much of the final translation rate can be determined by the TAAI.
The fact that codons affect the expression is well-known.1,2,8,51,52 Then we wonder whether the adaption of codon usage to isoacceptor tRNA gene copy number resulted in co-adaption between amino acid composition and tRNA gene copy number. We computed the correlation index according to codon usage of multi-organisms from three domains and multi-genes, and detected very weak adaption for codon usage to important translation rate-limiting factor tRNA copy number (Supplementary Table S6, Supplementary Fig. S1B, S3A, S3B). On the contrary of codon effects, the amino acid compositions should suffer the translation selection independently.
However, more and more researchers found that amino acid composition is under more translation selection than codon usage. Li et al. firstly reported that the codons decoded by rare transfer RNAs did not lead to slow translation under nutrient-rich conditions.17 Then, Subramaniam et al. showed that protein synthesis rates were highly similar across yellow fluorescent protein variants during amino acid-rich growth, while some synonymous codons were highly sensitive to environmental perturbation during limitation for corresponding amino acids.53 Zhou et al.54 lately reported that impacts of codon usage are mainly due to their effects on transcription and largely independent of translation. Williford and Demuth compared several expression related factors and introduced that highly expressed genes have stronger selection for amino acid composition than codon usage.55 It is proved that the cost minimization principle causes the amino acid relative abundances to connect with metabolic cost.56 In contrast to the consistency between the local effects of synonymous codon usage and tRNA gene copy number on translation rates,17,52 the TAAI is a translation rate associated factors with universal and global effects.
4.2. Energy efficiency and selective pressure for co-adaption
Co-adaption arises from energy efficiency and selective pressure. Organisms evolve to maximize efficiency and minimize energy cost by adapting through genetic mechanisms.40,49 Such global co-adaption might raise the translation efficiency globally, coinciding with the energy efficiency/ecological dynamics principle.50 Previously, Higgs and Ran analyzed 80 bacterial genomes and found that tRNA gene copy numbers evolved in response to translational selection.57 It is notable that they observed consistency between synonymous codon usage and tRNA gene copy number and that the unequal usage of synonymous codons encoding the same amino acid was involved. However, our current study on co-adaption focuses on disequilibrium frequencies among the twenty standard amino acids. Here, the co-adaption reflects a balance between tRNA gene copy number and the amino acids needed by the proteome. Redundant excessive tRNA gene copies will ultimately be a waste of translation resources ( did not increase the EGFP production in E. coli). Selective pressure drives the co-adaption at the protein level. Experiments showed that increasing the TAAI could indeed improve the translation speed of the proteins and hence validate that the co-adaption is caused directly by translation pressure. These results indicate that translation selection causes co-variation at the scale of organisms and individual proteins.
If we expand our view to the domains of life, which have evolutionary connections58,59, such selection also exists. We found that eukaryotic genomes had much better adaption values than the other two domains (Fig. 5). The translation rate for eukaryotic genomes is ∼3–8 amino acids per second,60 compared to 10–20 amino acids per second for bacterial genomes.61 In contrast, eukaryotes have much larger genomes and, hence, many more proteins. Thus, eukaryotic proteins would undergo stricter translation selective pressure. This higher pressure may be one of the reasons for the higher co-adaption observed in eukaryotes. Focusing on the bacterial domain, we observed that larger bacterial genomes tended to have higher TAAIs, and the TAAI value correlated positively with the genome size. Higher selective pressure may be the reason for this positive correlation. Taking all of the results into account, co-adaption is one effect of translation selection at all three levels (domain, genome and protein) and the selection complies with the maximum efficiency and minimum cost principle.
Figure 5.
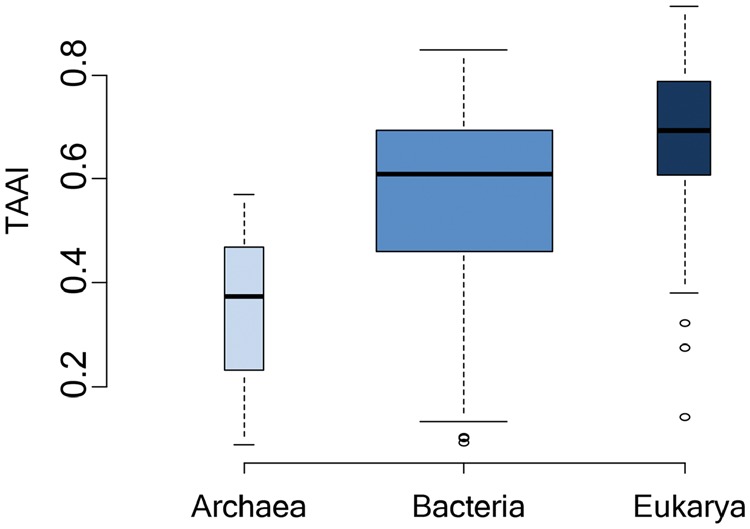
Comparison of the co-adaption (TAAI) of three domains. The medians for Archaea, Bacteria and Eukarya were 0.37, 0.61 and 0.69. Archaea had relatively low TAAIs, and Eukarya had the highest. Analysis of variance revealed that the three domains were significantly different, with a P value close to zero (P = 2.57e−06).
4.3. Potential application in lifting translation rate and hence protein production
This co-adaption can be applied to enhance translation efficiency in practice. Traditionally, in industrial application, the yield of a specific protein is improved by optimizing its synonymous codon usage.62 A higher ratio of optimal codons could facilitate the transcription efficiency by frequent usage of abundant or efficient tRNAs.14 Here, the yield of EGFP was improved markedly in E. coli by optimizing TAAI through increasing the gene copy number of specific tRNAs, thus increasing the translation speed at least tenfold. This finding may be applied in industrial production. To obtain higher output of one protein, we could optimize its co-adaption between tRNA gene copy number and amino acid usage by adding specific tRNA gene copies. Thus, the protein’s translation could be accelerated quickly. One of the prominent advantages of such an operation is that the yields of multiple proteins could be improved in one round. The production of multiple proteins could be increased by adding specific tRNA gene copies corresponding to their amino acid usage. This ideal result is based on the supposition that adding a specific tRNA gene could increase the TAAIs of many proteins simultaneously. A more practical method would be to divide all target proteins into groups based on similar amino acid frequencies. If the tRNA genes to import are carefully chosen, the target group of proteins will have higher expression levels but the other proteins should remain almost unchanged.
In the field of synthetic biology, it is hoped to devise and construct a general bacterial chassis cell that integrates functional synthetic parts, devices and systems. In practice, such a chassis has often been constructed or synthesized based on small and slowly growing bacteria.63,64 However, slow growth may limit their capacity to produce enough target molecules in a short time. Our strategy of importing certain tRNA genes may help to address this problem when designing chassis cells.
Supplementary Material
Acknowledgements
We thank Mr. Lu-Wen Ning for suggestions, Juan Feng’s lab and Lixia Tang’s lab for providing help with experimental materials and methods, and our lab members for fruitful discussion.
Supplementary data
Supplementary data are available at DNARES online.
Funding
This work was supported by the National Natural Science Foundation of China (31501063) and the Fundamental Research Funds for the Central Universities of China (ZYGX2016J117, ZYGX2015J144).
Conflict of interest
None declared.
References
- 1. Gingold H., Pilpel Y.. 2011, Determinants of translation efficiency and accuracy, Mol. Sys. Biol., 7, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang J.-R., Chen X., Zhang J.. 2014, Codon-by-codon modulation of translational speed and accuracy via mRNA folding, PLoS Biol., 12, e1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dekel E., Alon U.. 2005, Optimality and evolutionary tuning of the expression level of a protein, Nature, 436, 588–92. [DOI] [PubMed] [Google Scholar]
- 4. Sharp P. M., Li W.-H.. 1987, The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications, Nucleic Acids Res., 15, 1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. dos Reis M., Savva R., Wernisch L.. 2004, Solving the riddle of codon usage preferences: a test for translational selection, Nucleic Acids Res., 32, 5036–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ingolia N.T. 2016, Ribosome footprint profiling of translation throughout the genome, Cell, 165, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan X., Hoek T.A., Vale R.D., Tanenbaum M.E.. 2016, Dynamics of translation of single mRNA molecules in vivo, Cell, 165, 976–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fredrick K., Ibba M.. 2010, How the sequence of a gene can tune its translation, Cell, 141, 227–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tuller T., Carmi A., Vestsigian K., et al. 2010, An evolutionarily conserved mechanism for controlling the efficiency of protein translation, Cell, 141, 344–54. [DOI] [PubMed] [Google Scholar]
- 10. Gamble C. E., Brule C. E., Dean K.M., Fields S., Grayhack E.J.. 2016, Adjacent codons act in concert to modulate translation efficiency in yeast, Cell, 166, 679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elf J., Nilsson D., Tenson T., Ehrenberg M.. 2003, Selective charging of tRNA isoacceptors explains patterns of codon usage, Science, 300, 1718–22. [DOI] [PubMed] [Google Scholar]
- 12. Chan P.P., Lowe T.M.. 2009, GtRNAdb: a database of transfer RNA genes detected in genomic sequence, Nucleic Acids Res., 37, D93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nedialkova D.D., Leidel S.A.. 2015, Optimization of codon translation rates via tRNA modifications maintains proteome integrity, Cell, 161, 1606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cannarozzi G., Schraudolph N. N., Faty M., et al. 2010, A role for codon order in translation dynamics, Cell, 141, 355–67. [DOI] [PubMed] [Google Scholar]
- 15. Dittmar K.A., Goodenbour J.M., Pan T.. 2006, Tissue-specific differences in human transfer RNA expression, PLoS Genet., 2, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qu X., Wen J.-D., Lancaster L., Noller H.F., Bustamante C., Tinoco I.. 2011, The ribosome uses two active mechanisms to unwind messenger RNA during translation, Nature, 475, 118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li G.-W., Oh E., Weissman J.S.. 2012, The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria, Nature, 484, 538–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B.. 2013, Rate-limiting steps in yeast protein translation, Cell, 153, 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu C.-H., Dang Y., Zhou Z., et al. 2015, Codon usage influences the local rate of translation elongation to regulate co-translational protein folding, Mol. Cell, 59, 744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tuller T., Waldman Y.Y., Kupiec M., Ruppin E.. 2010, Translation efficiency is determined by both codon bias and folding energy, Proc. Natl. Acad. Sci. USA, 107, 3645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vargas-Rodriguez O., Musier-Forsyth K.. 2014, Structural biology: Wobble puts RNA on target, Nature, 510, 480–81. [DOI] [PubMed] [Google Scholar]
- 22. Subramaniam A.R., Zid B.M., O’Shea E.K.. 2014, An integrated approach reveals regulatory controls on bacterial translation elongation, Cell, 159, 1200–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. 2016, New structural insights into translational miscoding. Trends Biochemi. Sci., 41, 798–814. [DOI] [PubMed] [Google Scholar]
- 24. Espah Borujeni A., Salis H.M.. 2016, Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism, J. Am. Chem. Soc, 138, 7016. [DOI] [PubMed] [Google Scholar]
- 25. Wu B., Eliscovich C., Yoon Y.J., Singer R.H.. 2016, Translation dynamics of single mRNAs in live cells and neurons, Science, 352, 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mandel M.J., Silhavy T.J.. 2005, Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability, J. Bacteriol., 187, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ausubel F., Brent R., Kingston R., et al. 1987, Current protocols in molecular biology New York, New York: Wiley. [Google Scholar]
- 28. Benson D.A., Cavanaugh M., Clark K., et al. 2013, GenBank, Nucleic Acids Res., 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lowe T.M., Eddy S.R.. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. 2009, tRNAdb 2009: compilation of tRNA sequences and tRNA genes, Nucleic Acids Res., 37, D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang M., Weiss M., Simonovic M., et al. 2012, PaxDb, a database of protein abundance averages across all three domains of life, Mol. Cell. Proteomics, 11, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma L., Cui P., Zhu J., Zhang Z., Zhang Z.. 2014, Translational selection in human: more pronounced in housekeeping genes, Biol. Direct, 9, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yin H., Ma L., Wang G., Li M., Zhang Z.. 2016, Old genes experience stronger translational selection than young genes, Gene, 590, 29–34. [DOI] [PubMed] [Google Scholar]
- 34. Wei W., Jin Y. T., Du M. Z., Wang J., Rao N., Guo F. B.. 2016, Genomic complexity places less restrictions on the evolution of young coexpression networks than protein–protein interactions, Genome Biol. Evol., 8, 2624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garciavallve S., Guzman E., Montero M.A., Romeu A.. 2003, HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes, Nucleic Acids Res., 31, 187–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rocha E.P. 2004, Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization, Genome Res., 14, 2279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamao F., Andachi Y., Muto A., Ikemura T., Osawa S.. 1991, Levels of tRNAs in bacterial cells as affected by amino acid usage in proteins, Nucleic acids Res., 19, 6119–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levitt M. 2009, Nature of the protein universe, Proc. Natl. Acad. Sci. USA, 106, 11079–11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kempes C.P., Dutkiewicz S., Follows M.J.. 2012, Growth, metabolic partitioning, and the size of microorganisms, Proc. Natl. Acad. Sci. USA, 109, 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kempes C.P., Wang L., Amend J.P., Doyle J., Hoehler T.. 2016, Evolutionary tradeoffs in cellular composition across diverse bacteria, ISME J., 1010, 21452145–21572157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dana A., Tuller T.. 2014, The effect of tRNA levels on decoding times of mRNA codons, Nucleic Acids Res., 42, 9171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jost M.C., Hillis D.M., Lu Y., Kyle J.W., Fozzard H.A., Zakon H.H.. 2008, Toxin-resistant sodium channels: parallel adaptive evolution across a complete gene family, Mol. Biol. Evol., 25, 1016–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hale C.A., de Boer P.A.. 1997, Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell, 88, 175–85. [DOI] [PubMed] [Google Scholar]
- 44. Chang C., Stewart R.C.. 1998, The two-component system, Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol., 117, 723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kunkel T.A., Erie D.A.. 2005, DNA mismatch repair, Annu. Rev. Biochem., 74, 681–710. [DOI] [PubMed] [Google Scholar]
- 46. Titgemeyer F., Hillen W.. 2002, Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek, 82, 59–71. [PubMed] [Google Scholar]
- 47. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. 2009, Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling, Science, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Acosta-Rivero N., Sanchez J.C., Morales J.. 2002, Improvement of human interferon HUIFNalpha2 and HCV core protein expression levels in Escherichia coli but not of HUIFNalpha8 by using the tRNA(AGA/AGG). Biochem. Biophys. Res. Commun., 296, 1303–1309. [DOI] [PubMed] [Google Scholar]
- 49. Maitra A., Dill K.A.. 2015, Bacterial growth laws reflect the evolutionary importance of energy efficiency, Proc. Natl. Acad. Sci. USA, 112, 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grosskopf T., Consuegra J., Gaffe J., et al. 2016, Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long-term evolution experiment, BMC Evol. Biol., 16, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quax T.E., Claassens N.J., Söll D., van der Oost J.. 2015, Codon bias as a means to fine-tune gene expression, Mol. Cell, 59, 149–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duret L. 2000, tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes, Trends Genet., 16, 287–89. [DOI] [PubMed] [Google Scholar]
- 53. Subramaniam A.R., Pan T., Cluzel P.. 2013, Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acad. Sci. USA, 110, 2419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou Z., Dang Y., Zhou M., et al. 2016, Codon usage is an important determinant of gene expression levels largely through its effects on transcription, Proc. Natl. Acad. Sci. USA, 113, E6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Williford A., Demuth J. P.. 2012, Gene expression levels are correlated with synonymous codon usage, amino acid composition, and gene architecture in the red flour beetle, Tribolium castaneum, Mol. Biol. Evol., 29, 3755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krick T., Verstraete N., Alonso L.G., et al. 2014, Amino Acid metabolism conflicts with protein diversity, Mol. Biol. Evol., 31, 2905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Higgs P.G., Ran W.. 2008, Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage, Mol. Biol. Evol., 25, 2279–91. [DOI] [PubMed] [Google Scholar]
- 58. Booth A., Mariscal C., Doolittle W.F.. 2016, The modern synthesis in the light of microbial genomics, Annu. Rev. Microbiol., 70, 279–297. [DOI] [PubMed] [Google Scholar]
- 59. Lynch M., Conery J.S.. 2003, The origins of genome complexity, Science, 302, 1401–4. [DOI] [PubMed] [Google Scholar]
- 60. Mathews M.B., Sonenberg N., Hershey J.W.. 2000, Origins and principles of translational control, Cold Spring Harbor Monogr. Arch., 39, 1–31. [Google Scholar]
- 61. Liang S.-T., Xu Y.-C., Dennis P., Bremer H.. 2000, mRNA composition and control of bacterial gene expression, J. Bacteriol., 182, 3037–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Menzella H.G. 2011, Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli, Microb. Cell Fact., 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hutchison C.A. 3rd, Chuang R.Y., Noskov V.N., et al. 2016, Design and synthesis of a minimal bacterial genome, Science, 351, aad6253. [DOI] [PubMed] [Google Scholar]
- 64. Vickers C.E. 2016, The minimal genome comes of age, Nat. Biotechnol., 34, 623–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.