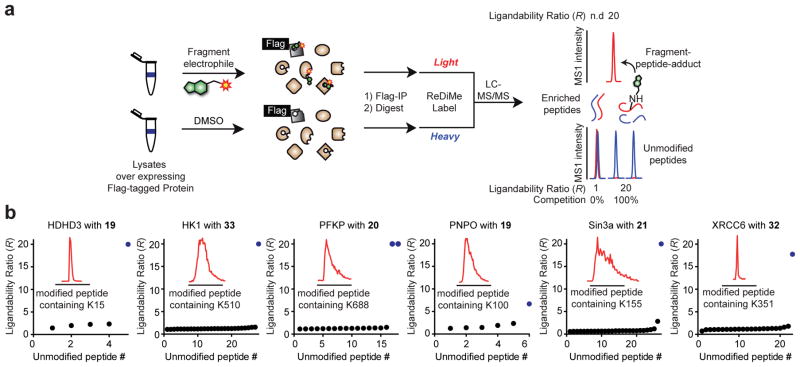
Figure 5.
Confirmation of site-specific fragment-lysine reactions by MS-based proteomics. a, Schematic workflow for direct measurement of lysine-fragment reactions on proteins by quantitative proteomics. Flag epitope-tagged proteins are recombinantly expressed in HEK 293T cells and the cellular lysates treated with DMSO or a fragment ligand and immunoprecipitated with anti-Flag agarose resin. The enriched proteins are eluted from the resin, digested with trypsin, and tryptic peptides from the DMSO and fragment-treated samples isotopically labeled by reductive dimethylation (ReDiMe) with heavy (blue) and light (red) formaldehyde40,41, respectively. The DMSO and fragment-treated samples are then combined and analyzed by LC-MS. Lysine-fragment reactions were confirmed by both: i) detection of the peptide-fragment adduct exclusively in the fragment-treated sample (top trace); and ii) depletion of the parent unlabeled tryptic peptides containing the indicated lysine or having the lysine at a proteolytic cleavage site (bottom trace). b, R values for all detected, unmodified lysine-containing tryptic peptides for representative liganded proteins after treatment with the indicated compounds at 50 μM for 1 h. Unmodified peptides that contain the liganded lysine or have it at a proteolytic cleavage site are shown as blue dots. MS1 chromatographic peaks for fragment-peptide adducts are shown in the inset traces.