Extended Data Figure E4. Base editing efficiencies of additional ABE2 and ABE3 variants, and the effect of adding A142N to TadA*–dCas9 on antibiotic selection survival in E. coli.
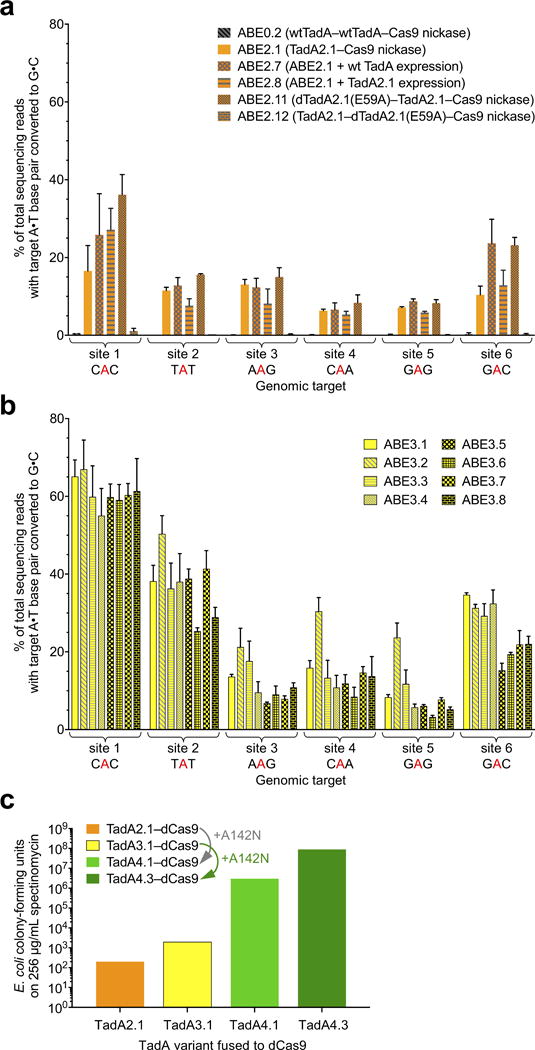
a, A•T to G•C base editing efficiencies in HEK293T cells at six human genomic target DNA sites of ABE2 variants with different engineered dimeric states. A control ABE variant containing two wild-type TadA domains and no evolved TadA* domains (ABE0.2) did not result in A•T to G•C editing at the six genomic sites tested, confirming that dimerization alone is insufficient to mediate ABE activity. b, A•T to G•C base editing efficiencies in HEK293T cells at six human genomic target DNA sites of ABE3.1 variants differing in their dimeric state (homodimer of TadA*–TadA*–Cas9 nickase, or heterodimer of wild-type TadA–TadA*–Cas9 nickase), in the length of the TadA–TadA linker, and in the length of the TadA–Cas9 nickase linker. See Extended Data Figure E1 for ABE genotypes and architectures. c, Colony-forming units on 2xYT agar with 256 μg/mL of spectinomycin of E. coli cells expressing an sgRNA targeting the I89T defect in the spectinomycin resistance gene and a TadA*-dCas9 editor lacking or containing the A142N mutation identified in evolution round 4. Successful A•T to G•C base editing at the target site restores spectinomycin resistance. Values and error bars in (a) and (b) reflect the mean and s.d. of three independent biological replicates performed on different days.
