Extended Data Figure E9. Analysis of cellular RNAs in ABE7.10-treated cells compared with untreated cells.
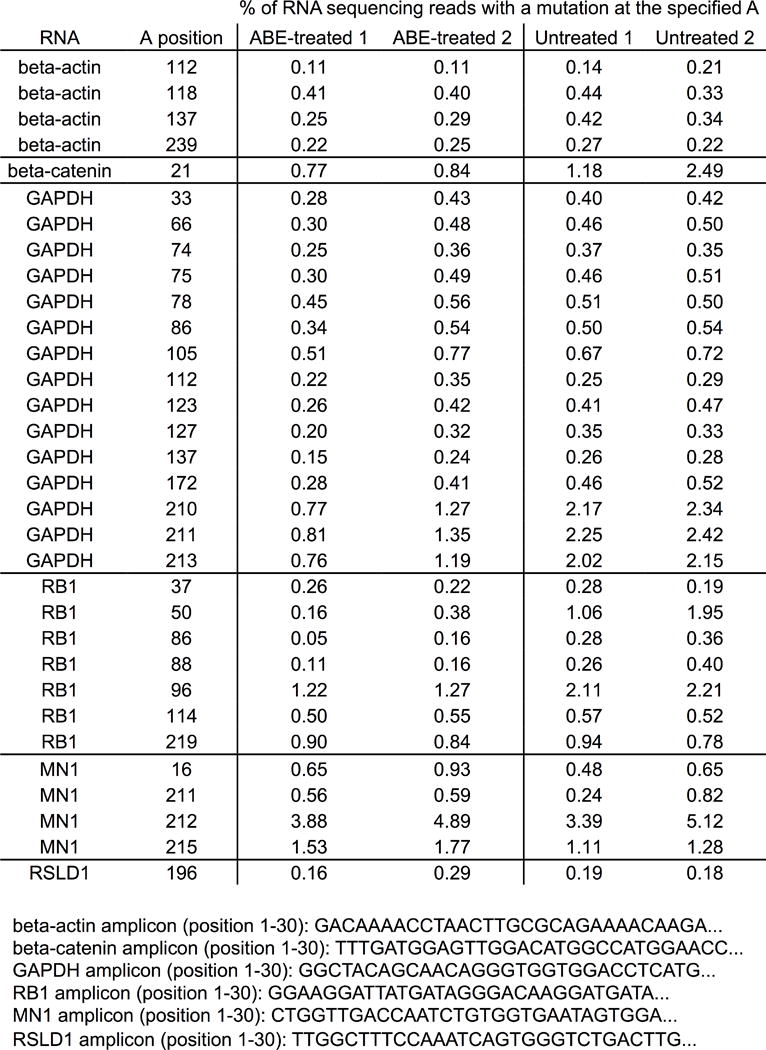
RNA from HEK293T cells treated with ABE7.10 and the sgRNA targeting site 1, or from untreated HEK293T cells, was isolated and reverse transcribed into cDNA. The cDNAs corresponding to four abundant cellular RNAs (encoding beta-actin, beta-catenin, GAPDH, and RB1), and two cellular RNAs with sequence homology to the tRNA anticodon loop that is the native substrate of E. coli TadA (encoding MN1 and RSLD1), were amplified and analyzed by HTS. Within each amplicon, the mutation frequency at each adenine position for which the mutation rate was ≥ 0.2% is shown for two ABE7.10-treated biological replicates (ABE-treated 1 and ABE-treated 2) and for two untreated biological replicates (untreated 1 and untreated 2). The start of each amplicon is shown.
