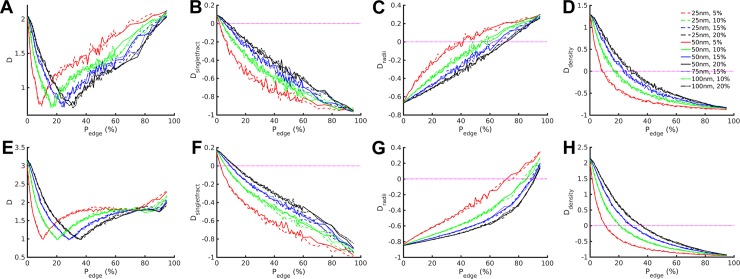
Fig 4. Model/Experiment discrepancy calculations quantify model performance and identify model parameterizations with optimum fit to experimental data.
To describe the goodness of fit of the models, we measured discrepancies of various simulation outputs as a function of Pedge. Data are displayed for untreated (A-D) and caspofungin-treated (E-H) yeasts. Note that Pedge = 0 corresponds to the random unmasking model. For each series, results are given for the total discrepancy (A,E) and the individual signed contributions of the three variables of interest [singlet fraction (B,F), equivalent radii of multi-exposures (C,G), and glucan exposure density (D,H)]. Dotted magenta lines in B-D and F-H denote the zero value for the indicated discrepancy metric, which is the value of optimum fit of model results to experimental results for an individual output. The above mean values are calculated from n = 100 runs for each condition.