FIG. 1.
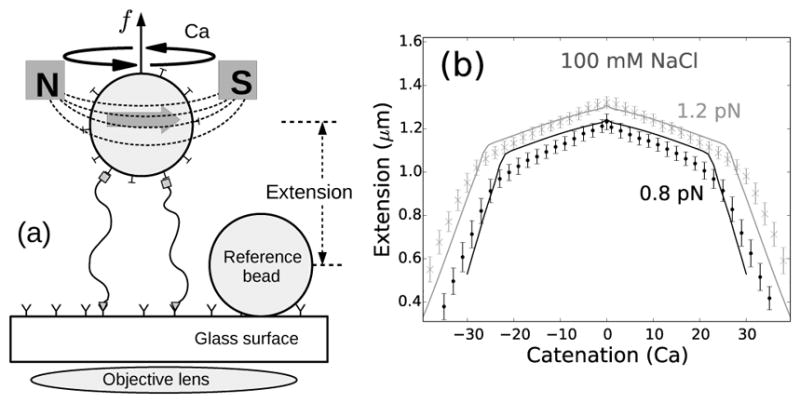
(a) Schematic of the magnetic tweezers setup. Two DNA molecules are attached (using only one of the DNA strands as attachments) to the glass surface and a paramagnetic bead via digoxigenin-antidigoxigenin and biotin-streptavidin interactions respectively. The single-strand attachments ensure no twisting of individual DNA molecules upon rotation of the bead [8, 23]. (b) Experimentally measured end-to-end extension of a braid as a function of catenation number for 0.8 (black points) and 1.2 pN (gray cross marks) applied forces in 100 mM NaCl, where the errorbars represent standard deviation. The solid lines represent theoretical predictions for 1.6 μm DNA molecules, with intertether distance 0.19 μm, and under 0.8 (black line) and 1.2 pN (gray line) force at 100 mM monovalent salt (Supplementary Material). The small peak in extension at zero catenation is due to the small intertether distance, i.e., the close proximity of the two DNA molecules for this particular DNA pair [22].
