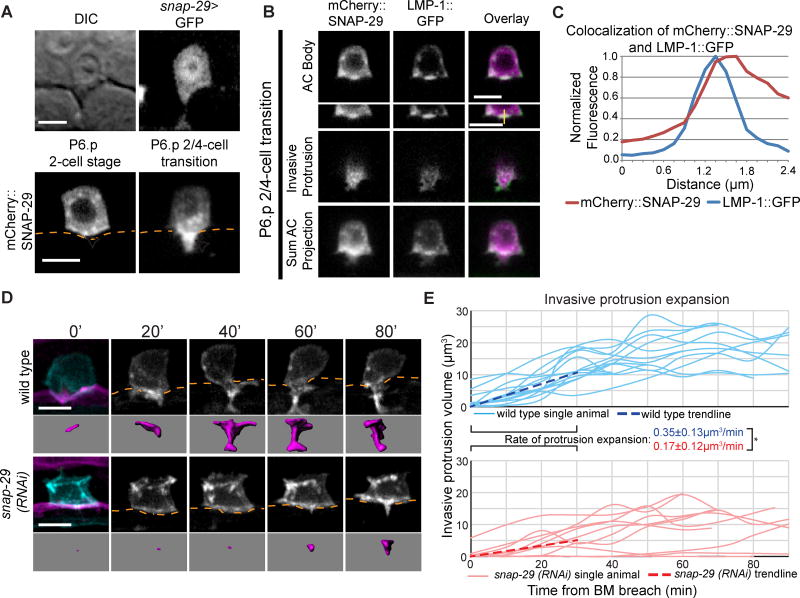
Figure 3. The t-SNARE SNAP-29 facilitates invasive protrusion expansion.
(A) A snap-29 transcriptional reporter (snap-29>GFP; top right) expressed in the AC. An AC expressed translational reporter (cdh-3>mCherry::SNAP-29; bottom) is polarized (arrowheads) to the invasive membrane before invasion (left) and within the protrusion (right); BM position, orange. (B) Colocalization of mCherry::SNAP-29 and LMP-1::GFP during protrusion formation. 0.5µm confocal z-slices of the AC body (top,) and invasive protrusion (middle); sum projection of the AC (bottom). Yellow line (top inset, right) indicates point of fluorescence measurement shown in (C). (C) Fluorescence measurement shows colocalization of peak SNAP-29 (red) and LMP-1 (blue) signal. (D) Time-lapse of AC protrusion formation in a uterine specific RNAi strain. AC membrane labeled by cdh-3>mCherry::PLCδPH, blue and greyscale, purple isosurface below; BM (laminin::GFP, magenta) position shown by orange dotted lines. (E) AC protrusion volume over time for wild type (blue) and snap-29 (RNAi) (red) animals. Average expansion rates (dotted lines) ± SD for first 30 minutes of protrusion growth (n=14 animals for control, n=10 animals for snap-29 (RNAi); *p<0.05, Student's t-test). Wild type controls were the same as in Figure 2D. Scale bars = 5µm. See also Figure S3.