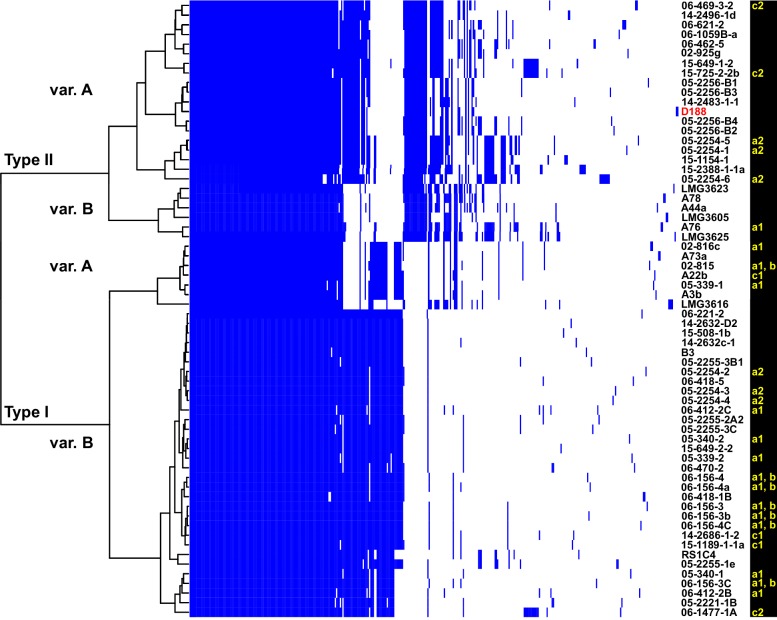
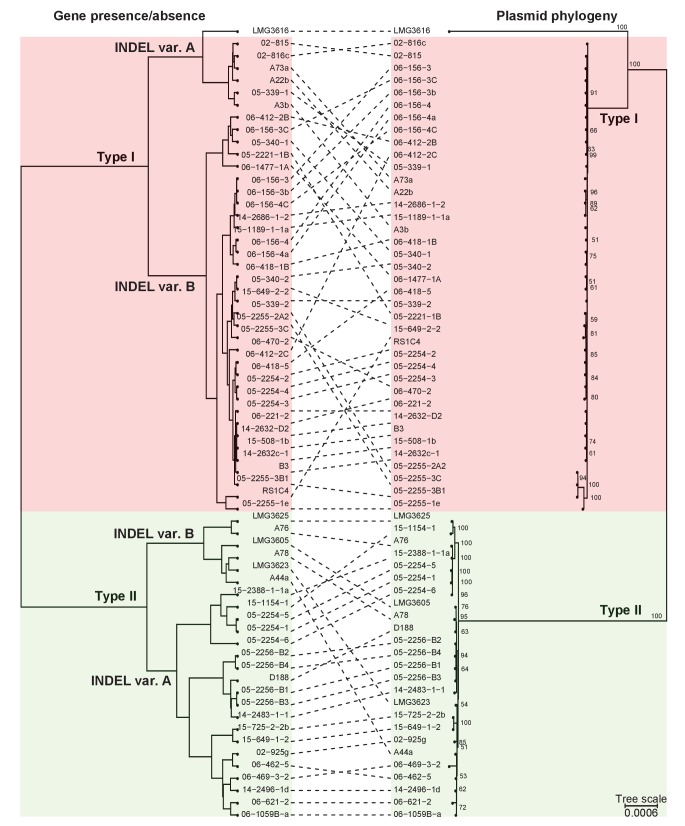
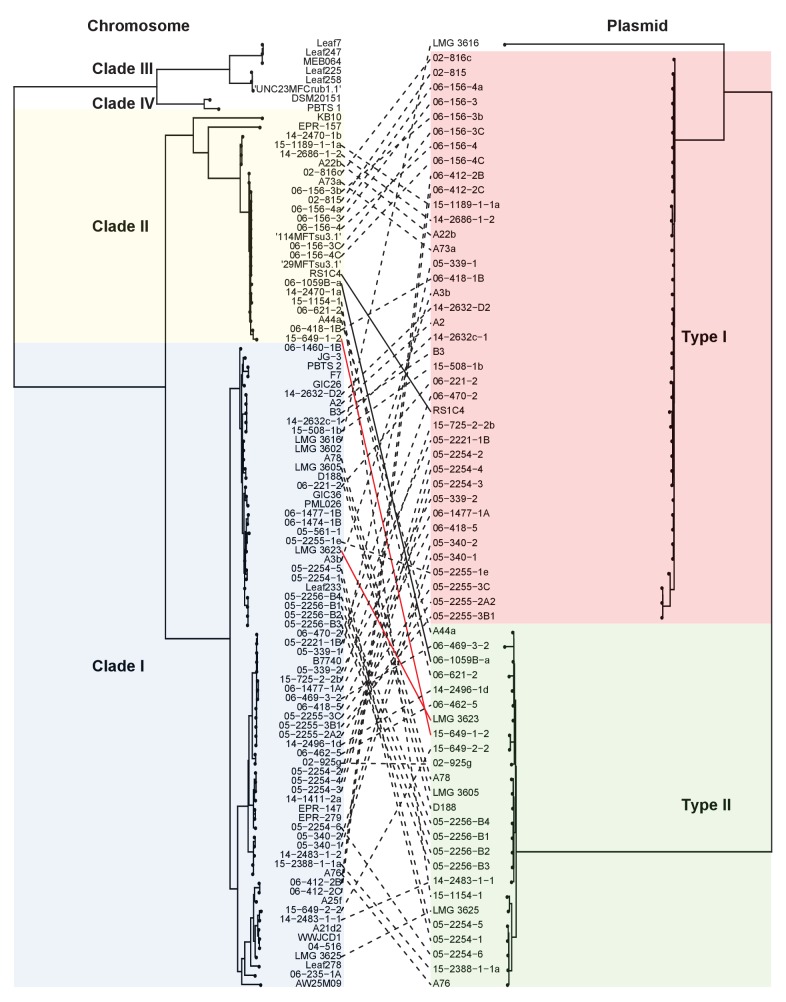
Figure 3. Analysis of plasmid variation reveals multiple patterns in distribution.
Rows indicate genes present in (blue) or absent from (white) the plasmids of isolates (listed to the right; D188 is labeled in red for reference). Columns represent individual genes. Type categories were determined on the basis of phylogenetic analysis of the core genes. INDEL variants, delineated by gray and white shading, were determined on the basis of the cladogram. The plasmid carried by LMG3616 is enclosed by dotted lines. The lower-case letters and numbers (a1, a2, b, c1, and c2) listed along the right, relate isolates and their plasmids to the potential transmission patterns indicated in Figure 2.