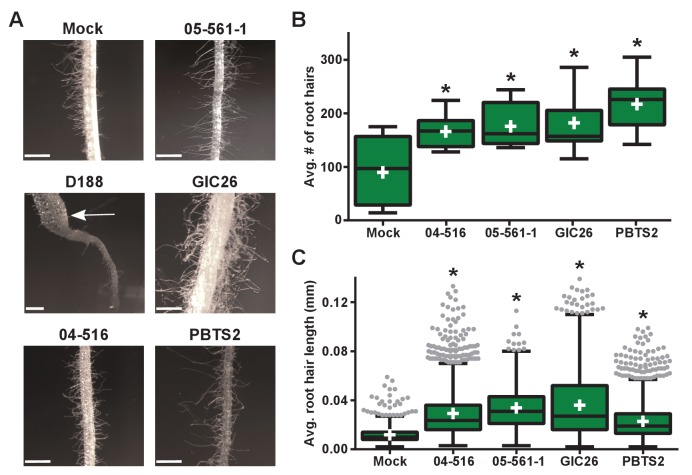
Figure 5. Plant-associated Rhodococcus bacteria cause changes to the root architecture of seedlings.
(A) Representative images of root hairs of N. benthamiana that were inoculated with isolates of Rhodococcus. Images were taken 25 days post inoculation (dpi). The white arrow indicates the thicker stem induced only by isolate D188. Scale bars are 0.5 mm. (B) Quantification of average root hair number at 25 dpi. All root hairs were manually counted for at least five seedlings per treatment. (C) Quantification of root hair lengths at 25 dpi. All root hairs were manually measured for at least five seedlings per treatment. For B and C, data were repeated in two independent biological replicates. * indicates a significant difference compared to the mock treatment.
Figure 5—figure supplement 1. Five additional virulence-gene-lacking isolates of Rhodococcus cause changes to root architecture.