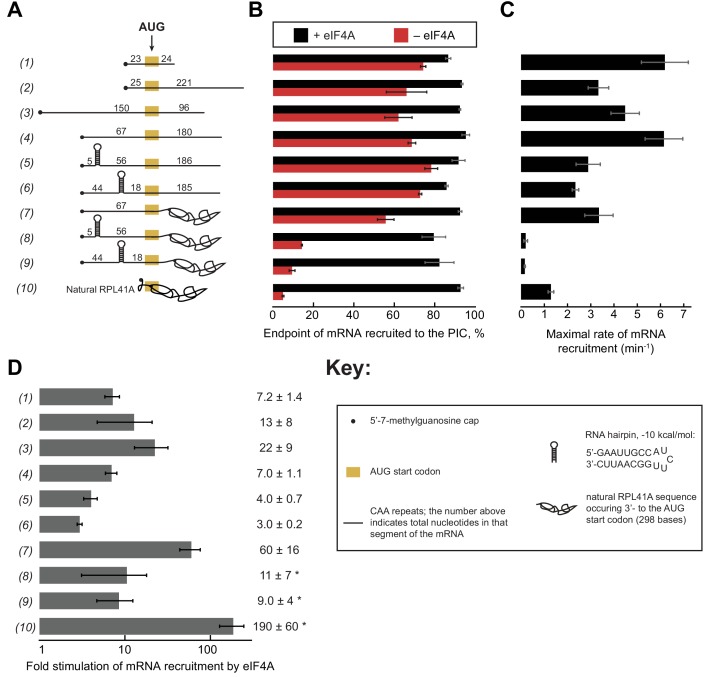
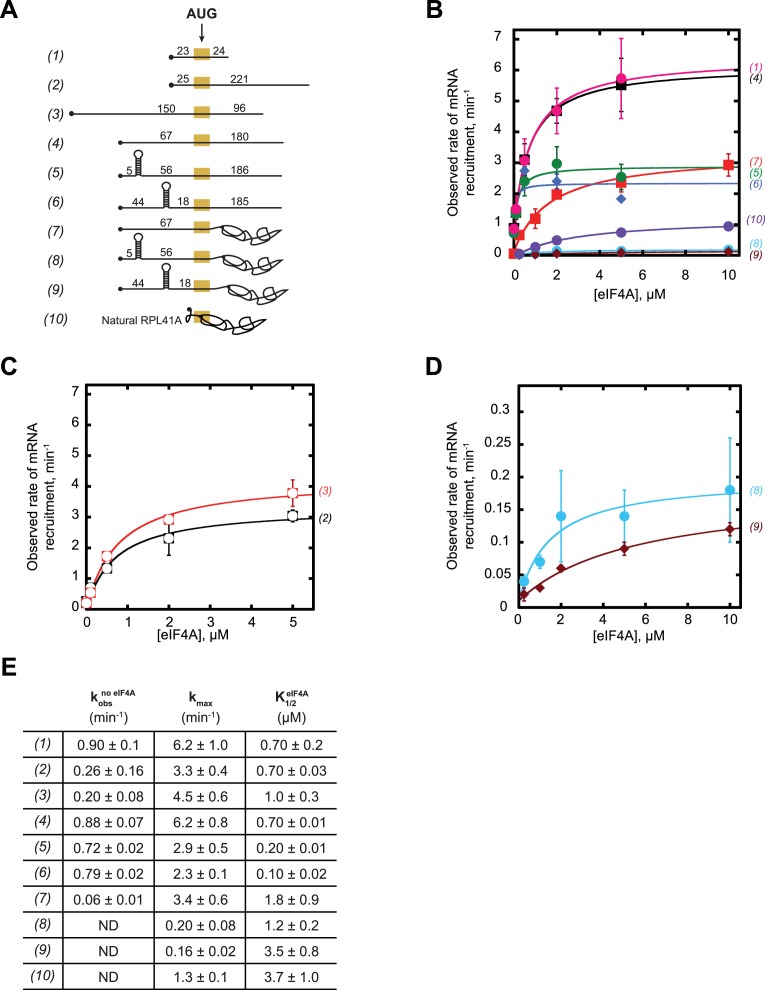
Figure 3. eIF4A stimulates recruitment of mRNAs regardless of their degree of structure.
(A) Schematic of mRNAs used in the study. mRNAs were capped unless otherwise noted but do not contain a poly(A) tail. Numbers on the mRNA indicate the total number of nucleotides in the corresponding segment of the RNA. (B) Endpoints of recruitment to the PIC for mRNAs in (A) in the presence (black) or absence (red) of saturating eIF4A. mRNAs are listed in the same order as in (A). (C) Maximal rates of mRNA recruitment, kmax (min−1), measured for the mRNAs in (A) (the corresponding plots are shown in Figure 3—figure supplement 1B–D) listed in the same order as in (A). (D) eIF4A-dependent stimulation of mRNA recruitment: the maximal rate of mRNA recruitment at saturating eIF4A concentration divided by the observed rate in the absence of eIF4A (calculated from data in Figure 3—figure supplement 1E). Numbers to the left correspond to the mRNAs in (A). The asterisk (*) indicates that due to low recruitment endpoints in the absence of eIF4A, data for mRNAs 8–10 could not be fit with an exponential rate equation and thus the fold stimulation by eIF4A was estimated from comparison of initial rates in the presence of saturating eIF4A versus the absence of eIF4A. All data presented in the figure are mean values (n ≥ 2) and error bars represent average deviation of the mean.