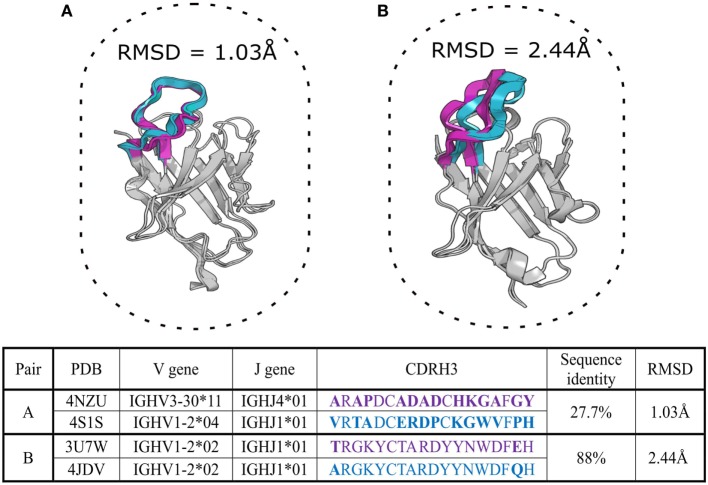
Figure 2.
Two aligned pairs of VH chains extracted from SAbDab, the antibody structural database (29). Complementarity determining region-3 of the heavy chain (CDR-H3) sequences in pair (A) belong to different CDR-H3 clonotypes but adopt very similar structural configurations with a root mean square deviation (RMSD) of ~1 Å. Pair (B) includes germline precursor (4JDV) and matured (3U7W) anti-gp120 antibodies (78, 79). Although CDR-H3 sequences of pair (B) are members of the same clonotype, the RMSD shows that their CDR-H3 shapes are structurally distinct (RMSD > 2 Å). CDR-H3 loops and their amino-acid sequences are in purple and cyan colors, mismatched amino acid are in bold. The RMSD of the backbone atom positions of proteins provides a pairwise measurement of the three-dimensional dissimilarity between two sets of coordinates where solved or predicted structures are available. Sub-Angstrom RMSD indicates structurally identical shapes, while an RMSD value greater than 2 Å for a short segment indicates structurally distinct configurations (80).