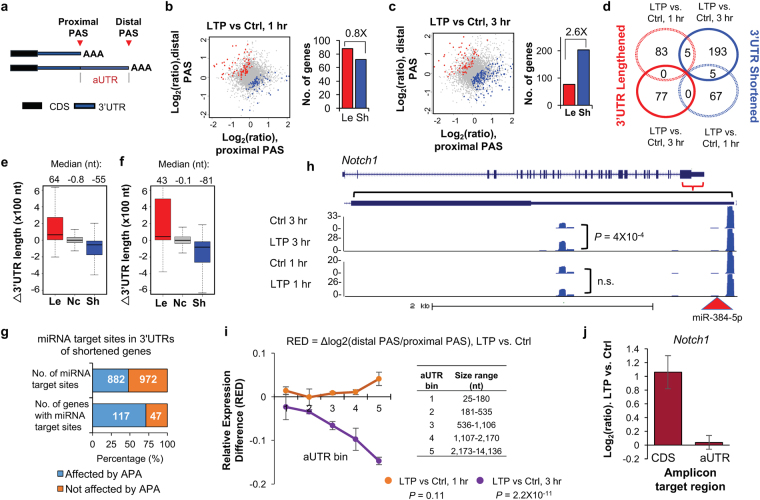
Figure 2.
Regulation of 3′UTR APA after LTP induction. (a) Schematic of 3′UTR APA. (b) 3′UTR APA regulation after 1 hr LTP induction. Left, Scatterplot comparing expression changes of proximal and distal PASs after 1 hr LTP induction. Genes that significantly switched to proximal PAS usage are in blue and those that switched to distal PAS usage are in red (P < 0.05, DEXSeq, and relative abundance change >5%). Grey dots are genes without significant APA regulation. Right, bar graph comparing the number of genes with lengthened or shortened 3′UTRs (Le and Sh, respectively). (c) As in (b), 3 hr post LTP induction. (d) Venn diagram comparing genes with significant 3′UTR regulation 1 hr post LTP induction and 3 hr post LTP induction. (e) 3′UTR length change in 1 hr post LTP induction. Genes with 3′UTRs significantly shortened (blue), lengthened (red), or unchanged (grey) are shown. 3′UTR size was based on weighted mean of all 3′UTR isoforms. Median values are indicated on the top. (f) As in (e), except that data are based on 3 hr post LTP induction. (g) Effect of 3′UTR shortening on miRNA targeting. Number of miRNA target sites that are removed (882) or not removed (972) by 3′UTR shortening is indicated in the upper bar, and number of genes that contain removed (117) or not removed (47) miRNA target sites by 3′UTR shortening are indicated in lower bar. (h) An example gene Notch1, which displayed 3′UTR shortening after LTP. UCSC genome browser tracks of 3′READS data are shown. Gene structure and 3′UTR sequence are indicated on top. Peaks from two polyA sites indicate reads for corresponding APA isoforms. miR-384-5p target site is indicated. Reads are based on combined samples. (i) Relationship between aUTR size and 3′UTR-APA regulation at 1 hr or 3 hr post LTP induction. Relative expression difference (RED) was calculated for all genes with APA sites. The formula of RED is indicated above the graph. Genes were divided into five groups (aUTR bins, listed on the right) based on aUTR size, with approximately equal number of genes in each bin. For each bin, mean RED was calculated. Error bars are standard error of mean (SEM). P-value (Wilcoxon rank sum test) indicates the difference between bin 1 and bin 5. (j) RT-qPCR validation of Notch1 APA regulation. Two different primer sets were used to detect the expression of a common coding region (left bar) and a region in the alternative 3′UTR (right bar). Error bars are SEM of 4 replicates.