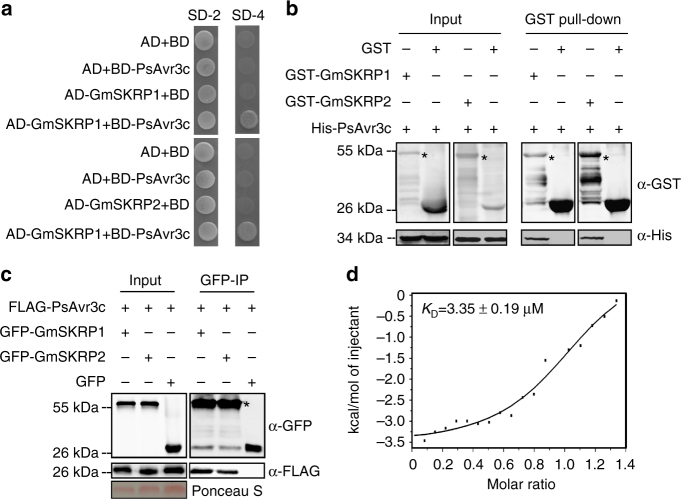
Fig. 2.
PsAvr3c physically interacts with soybean GmSKRP1/2 in vitro and in vivo. a PsAvr3c interacts with GmSKRP1/2 in yeast. PsAvr3c and GmSKRP1/2 were cloned into bait plasmid pGBKT7 (BD) and prey plasmid pGADT7 (AD), respectively. Yeast transformants were grown on SD/-Trp/-Leu (SD-2) and selected on SD/-Trp/-Leu/-His/-Ade (SD-4). The plates were photographed 5 days after inoculation. Experiments were repeated three times with similar results. b PsAvr3c physically interacts with GmSKRP1/2 in vitro. GST-GmSKRP1/2 or GST bound resins were incubated with E. coli supernatant containing His-PsAvr3c. Protein bands of GST-GmSKRP1/2 are marked by asterisks. The presence of His-tagged proteins was detected by western blot using anti-His tag antibody. c PsAvr3c interacts with GmSKRP1/2 in vivo. Co-immunoprecipitations (IP) were performed in extracts of N. benthamiana leaves expressing both FLAG-PsAvr3c and GFP-GmSKRP1/2. The presence of FLAG proteins was detected by western blot using anti-FLAG antibody. The GFP-GmSKRPs protein bands are indicated by asterisks and the protein loading is indicated by Ponceau stain. d Measuring binding affinity between PsAvr3c and GmSKRP1 in vitro. The binding affinity was determined using isothermal titration calorimetry (ITC). The raw titration data were integrated and fitted to a one-site binding model using the MicroCal PEAQ-ITC analysis software. Three original measurement results are 3.38, 3.52 and 3.14 μM, K D = 3.35 μM is mean value of three independent ITC experiments