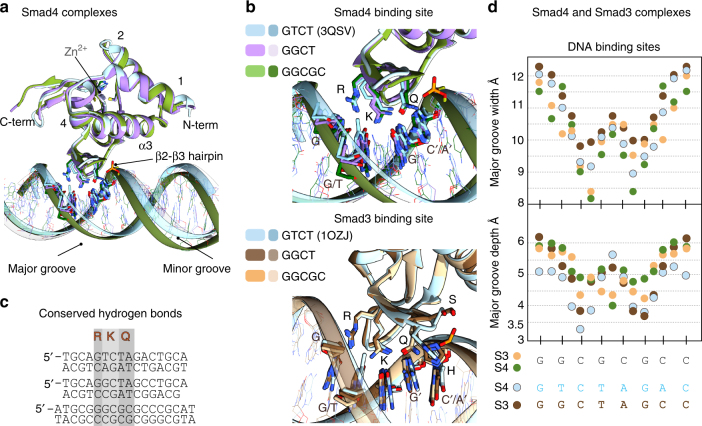
Fig. 6.
Consensus binding site for GC motifs and DNA shape comparison. a Superposition of the human Smad4 complexes with the GGCGC site (shown in green) and the GGCT site (in violet) to the GTCT structure (sky blue) previously determined (PDB: 3QSV). b Superposition of Smad4 (Top) and Smad3 (bottom) binding sites. Nucleotides that interact with the protein are highlighted. The Smad4 complexes are colored as in a. Smad3 complexes are displayed in tan (GGCGC site), brown (GGCT), and light blue GTCT (structure previously determined PDB:1OZJ). c Structural alignment of the DNA-binding sites observed for the six complexes, with the contacts to the three conserved residues R, K, and Q highlighted. d DNA shape comparison for different complexes. Major groove width (top) and depth (bottom) were calculated using Curves+ for Smad3 MH1–GGCGC complex (tan), Smad3 MH1–GGCT (brown), Smad4 MH1–GGCGC complex (green), and Smad4 MH1 (light blue) bound to the GTCT site (PDB: 3QSV). The analyzed sequences are indicated