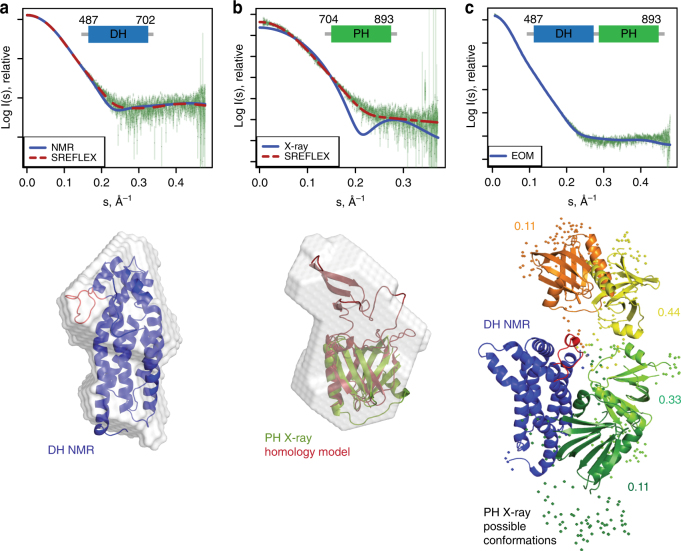
Fig. 3.
Small-angle X-ray scattering (SAXS) of the DH–PH tandem domain. a Experimental SAXS profile of the DH domain (green dots) and theoretical scattering profiles for the NMR model (discrepancy χ 2 = 1.5) and SREFLEX refined model (discrepancy χ 2 = 1.0). Below, the DH domain NMR structure is superimposed with the SAXS ab initio model as gray envelope. The α4–α5 loop is highlighted in red. b Experimental SAXS profile of the PH domain (green dots) and theoretical scattering profiles of PH crystal structure (blue, discrepancy χ 2 = 8.1), as well as a refined model for the full-length PH domain (red, discrepancy χ 2 = 1.1). Modeling of the full-length PH domain was based on a homology model with Xpln as a template (PDB ID 2Z0Q). For the PH domain, the SAXS based ab initio model is shown superimposed with the refined model for the full-length PH domain (red) and the PHΔ770–829 crystal structure (green). c Experimental SAXS profile of the DH–PH domain (green dots) and theoretical scattering of a representative solution ensemble after EOM (blue, discrepancy χ 2 = 1.0). Possible conformations of the DH–PH tandem domain are shown after superposition on the DH domain (blue). The respective orientations of the PH domain are represented in orange, yellow, light, and dark green, and the fractions for the respective PH domain positions are indicated next to the structures illustrating the inter-domain flexibility. Regions without high-resolution structural data, including the DH–PH linker and the internal PH deletion 770–829, are shown as spheres