Abstract
G-quadruplexes (G4) within oncogene promoters are considered to be promising anticancer targets. However, often they undergo complex structural rearrangements that preclude a precise description of the optimal target. Moreover, even when solved structures are available, they refer to the thermodynamically stable forms but little or no information is supplied about their complex multistep folding pathway. To shed light on this issue, we systematically followed the kinetic behavior of a G-rich sequence located within the c-KIT proximal promoter (kit2) in the presence of monovalent cations K+ and Na+. A very short-lived intermediate was observed to start the G4 folding process in both salt conditions. Subsequently, the two pathways diverge to produce distinct thermodynamically stable species (parallel and antiparallel G-quadruplex in K+ and Na+, respectively). Remarkably, in K+-containing solution a branched pathway is required to drive the wild type sequence to distribute between a monomeric and dimeric G-quadruplex. Our approach has allowed us to identify transient forms whose relative abundance is regulated by the environment; some of them were characterized by a half-life within the timescale of physiological DNA processing events and thus may represent possible unexpected targets for ligands recognition.
INTRODUCTION
Nucleic acids can fold into a variety of secondary structures (1). Among them, G-quadruplexes (G4), formed within G-rich DNA or RNA sequences, are of current interest. The basic unit of G4 is the G-tetrad, a square planar array of four guanines interacting through Hoogsteen hydrogen bonds. Two or more G-tetrads assemble through π stacking interactions to build the final G4 arrangement (2). G4 DNA may be comprised of mono-, bi- or tetra-molecular arrangements, although within genomic DNA unimolecular forms are expected to predominate. Overall G4 may also be clustered into parallel, antiparallel or mixed hybrid forms, as defined by participating strand orientation and loop topology.
Recent studies highlighted frequent G-quadruplex formation at oncogene promoters and suggested their relevant role in transcription regulation (3,4). Based on these observations, much effort has been directed toward the identification of small molecules able to modulate the conformational equilibria of these G-rich sequences in a specific and controlled way. The general starting point of modern drug design is the availability of a high-resolution structure of the target obtained by NMR and/or crystallographic techniques. These data provide essential information concerning the final thermodynamically stable structures assumed by the target sequences, but little or no information is supplied about their folding pathway. Nevertheless, oligonucleotide folding into a G4 tends to be a very complex, multistep pathway, and this can be critical since the final structure is achieved passing through different folding intermediates (5).
Another common feature of many G4s is that the thermodynamic equilibrium is reached very slowly. In contrast, recent studies showed that transcriptional processes are rapid events, considering that RNA polymerase II elongation rates in mammalian cells range between 1.3 and 4.3 kb/min (6,7). Additionally, transcription factors bind their specific sites on a time scale of seconds and their residence time varies from seconds to minutes (7–13). It can be concluded that in order to affect the binding of transcription factors to their consensus sequences and to stop the transcription, a G4 in the gene promoter should fold in a timescale close to the activity of the transcriptional machinery. This makes it clear that investigation of short-lived intermediates and folding rates are crucially important. Such short-lived structures might represent a suitable target for G4 ligands leading to unique downstream effects.
A good model for these studies is represented by kit2, a well-known G-rich sequence within the c-KIT gene promoter. This gene codes for a tyrosine–kinase receptor which, once activated, participates in a broad range of physiological processes, including cell proliferation, migration, maturation and survival (14,15). Recently, induction of G4 structures within this promoter has been associated with reduction of the expression of the receptor, possibly leading to important anti-cancer effects (16–18). Previous work highlighted the strong polymorphic behavior of kit2 (19,20). In fact, using proper mutants, two different G-quadruplex forms, a monomer and an intertwined dimer, have been resolved, both arranged according to a parallel conformation (21). In solution, the wild type sequence can assume both of these structures resulting in slow folding processes that progressively affect the population distribution (20). Once formed, the monomer and dimer seemingly do not interconvert on a fast time scale, suggesting that they might be derived from different folding pathways (22). All of this evidence suggests that a discrepancy between the solved structures and the pharmacological target(s) could actually exist.
The main purpose of this study was to dissect the G4 folding pathway of kit2 under different ionic conditions by following its kinetic behavior through the application of analytical ultracentrifugation, spectroscopic and electrophoretic techniques in order to derive information about the topology of intermediates that might be relevant during the physiological remodeling of the promoter occurring during the cell cycle. The resulting picture is expected to enable the design of selective ligands able to stop the transcription of c-KIT in a more targeted and efficient way.
MATERIALS AND METHODS
Materials
kit2 oligonucleotide 5′-CGGGCGGGCGCGAGGGAGGGG-3′ was purchased from Eurogentec (Liège, Belgium) that performed an RP-HPLC purification of the products. We checked the quality of the oligonucleotides by means of PAGE and HPLC without further purification. The oligonucleotide was dissolved in 10 mM Tris, pH 7.5, to prepare a 1 mM stock solution (strand concentration). Before use, each sample was heated at 95°C for 10 min in the required buffer and then slowly cooled down at room temperature to equilibrate the system.
Circular dichroism
Circular dichroism experiments were performed on a JASCO J-810 spectropolarimeter equipped with a Peltier temperature controller. Measurements were obtained using a 1 cm path-length quartz cuvette. CD spectra were recorded from 230 to 320 nm, with the following parameters: scanning speed 100 nm/min; band width of 2 nm; data interval of 0.5 nm; response of 2 s. The contribution of the buffer was subtracted from the sample spectra after each acquisition. The nucleic acid concentration, the salt concentration, the buffer composition and the temperature used for the experiments varied according to the purpose of the assays.
For CD kinetic experiments, the selected cation was added manually to the cuvette from a stock solution and mixing was provided by an in-cuvette magnetic stirring bar. After a mixing time of 5 seconds, spectra acquisition was initiated, using an interval scan of 60 s. Observed ellipticities were converted to molar ellipticity [Θ] which is equal to deg·cm2·dmol−1 (Mol. Ellip.) calculated using the DNA residue concentration in solution.
Polyacrylamide gel electrophoresis (PAGE)
For each sample, 200 ng of oligonucleotide were heated at 95°C for 10 min in 10 mM Tris pH 7.5 and allowed to cool to room temperature overnight. Afterward, increasing concentrations of KCl (0–50 mM) were added to the samples and they were left to equilibrate for 24 h at room temperature. Then the samples were loaded on a native 15% polyacrylamide (19:1 acrylamide: bisacrylamide) PAGE in 1× TBE. A scrambled oligonucleotide (M, which sequence is 5′-GGATGTGAGTGTGAGTGTGAGG-3′) of the same molecular weight of the studied sequence was used as an electrophoretic mobility marker. The gel was stained using SYBR green II, and the resolved bands were visualized on an image acquisition system (Geliance 600 Imaging system, Perkin-Elmer).
Analytical Ultracentifugation
AUC was carried out in a Backman Coulter ProteomeLab XL-A analytical ultracentrifuge (Beckman Coulter Inc., Brea, CA, USA) at 25°C overnight at 50 000 rpm in standard 2 sector cells. Each sample contained 2 μM kit2 in 10 mM Tris pH 7.5 equilibrated with/without 50 mM KCl or NaCl. Data were analysed using the program Sedfit (www.sedfit.com). The concentration-dependent distributions of sedimenting species were calculated using the c(S) continuous distribution model considering measured values for buffer density and viscosity. Buffer density was measured on a Mettler/Paar Calculating Density Meter DMA SSA at 20°C, and viscosity was measured using an Anton Paar AMVn Automated Microviscometer. For the calculation of frictional ratio, 0.55 ml/g was used for partial specific volume and 0.3 g/g was assumed for the amount of water bound.
Stopped-flow UV kinetic experiments
Fast folding steps were studied by using a UV stopped flow apparatus equipped with a rapid scanning monochromator (On-Line Instrument System, Borgat, GA, USA). Each sample was prepared in 10 mM Tris pH 7.5. DNA concentrations and cations were changed according to the purpose of the experiments. The UV-absorbance spectra were recorded from 270 to 330 nm with an acquisition rate of 1 ms/spectrum. A water bath maintained the system at a constant temperature of 25°C. Three mixing experiments were averaged for each data analysis. In order to avoid artifacts, buffer–buffer and buffer–oligonucleotide mixing experiments were performed as controls.
Data analysis
Single-wavelength kinetic experiments were analyzed by nonlinear least square using single or multiple exponential fitting functions.
The best fits for UV stopped flow experiments were obtained using a bi-exponential fitting function (Equation 1),
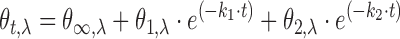 |
(1) |
The best fitting for single wavelength CD kinetic experiments were obtained by applying a model that considers concurrent, independent first-order and second order-kinetic processes. The terms describing these two processes (reported in Table 1 in Ref (23)) were linearly combined to derive Equation (2):
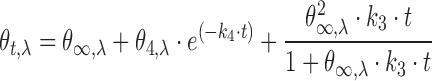 |
(2) |
where θt,λ is the value of signal at time t and θ∞,λ is the final value of the signal. θ1–2-4,λ are amplitude factors for each exponential, while k1–2-3–4 are kinetic constants.
Table 1. Distribution of the kit2 species in solution as derived by AUC analysis in 10 mM Tris pH 7.5 with/without 50 mM KCl and comparison of their experimentally determined sedimentation coefficients with those calculated by HydroPro software on solved structures.
| Species in solution | Distribution in solution | Sedimentation coefficient (S) | |
|---|---|---|---|
| HydroPro | AUC | ||
| Monomer | 60% | 1.50a | 1.52 |
| Dimer | 40% | 2.53b | 2.65 |
| Monomer (no KCl) | 100% | — | 1.47 |
aData calculated on PDB 2KYP.
bData calculated on PDB 2KYO.
Singular value decomposition (SVD)
Multiple wavelength CD kinetic experiments were analysed using SVD. This dataset consists of a matrix θi,j, where θi,j is the ellipticity at wavelength i and time j. The data matrix containing the time-dependent CD spectra, named D matrix, is broken up into three different sub-matrices: the S matrix, that keeps information about how much every single species contributes to the dichroic signal; the U matrix, that maintains data concerning the spectral shapes of all the conformations in solution; and the V matrix, that indicates how the spectral changes occur over time. The combination of S, U and V matrices provides the initial D matrix, so that D = U × S × V. The S values and U and V autocorrelation coefficients allow determination of the number of species that significantly contribute to the dichroic changes. The best fitting of the significant V eingenvectors was obtained by applying a model considering first order and second order kinetic processes (Equation 2) and determining the kinetic constants and other fitting parameters that contribute to the H matrix. By the linear combination of U × S matrix and H matrix, we obtained the actual spectra of the species in solution significantly contributing to the dichroic signal (24–26). Matlab, Olis Globalworks and Sigmaplot packages were used to perform SVD analysis.
RESULTS
Folded species distribution in potassium-containing solution
As highlighted in the introduction, kit2 can fold into different G-quadruplex structures with different kinetic parameters (20). Our first aim was to assess whether the major forms present in solution at equilibrium are consistent with the high-resolution structures obtained by NMR on mutated kit2 sequences.
We started the evaluation of the folding process of kit2 by considering the system at thermodynamic equilibrium. In order to monitor the relative distribution of the species in solution in this condition, PAGE experiments were performed following the folding of kit2 in the presence of increasing concentrations of KCl.
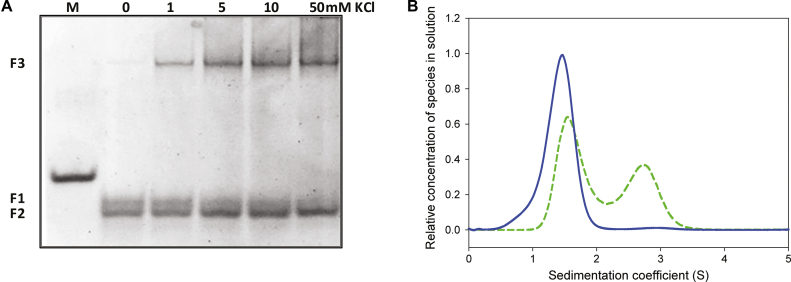
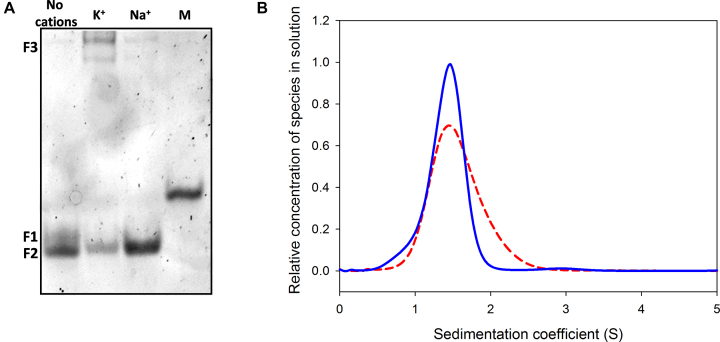
As shown in Figure 1A, in the absence of metal ion (lane 0), kit2 shows two different electrophoretic bands (F1 and F2), both with higher mobility when compared to a scrambled oligonucleotide of the same molecular weight (first lane, M). After addition of potassium, the upper band (F1) disappears and converts into F2 and into a new band (F3) with remarkably lower electrophoretic mobility. At 50 mM KCl, the conversion of F1 is complete. Previous data confirmed that these bands can be extracted from the gel with negligible interconversion, thus allowing one to acquire the corresponding CD spectra, both of which are compatible with a parallel G-quadruplex (22). Consistently with the involvement of the monovalent cation, we can assign F2 and F3 to monomeric and dimeric G-quadruplexes, respectively. These conclusions were confirmed by analytical ultracentrifugation (AUC) that allows one to determine the sedimentation coefficient of each species in solution (Figure 1B).
Figure 1.
(A) Electrophoretic resolution of kit2 in presence of increasing KCl concentration (0–50 mM) performed on 15% acrylamide native PAGE in 1× TBE. M refers to a scrambled oligonucleotide of the same molecular weight, (B) relative distribution of kit2 species in solution as function of their sedimentation coefficient with/without 50 mM KCl (respectively, dashed and solid lines) determined in 10 mM Tris pH 7.5.
AUC data showed that kit2 is mostly monomeric in the absence of monovalent cations (Figure 1B, solid line). In the presence of 50 mM KCl (Figure 1B, dashed line), the population of oligonucleotide splits into two groups, probably monomeric and dimeric forms. The experimentally determined sedimentation coefficients match those calculated using HydroPro software based on the reported NMR solved structures of the monomer and the dimer (PDB: 2KYP and PDB: 2KYO; respectively; Table 1). The integration of the obtained peaks showed that at the equilibrium 60% of kit2 in solution assumes the monomeric form whereas the remaining corresponds to the dimer.
It is important to remember that the dimeric form does not simply derived from the interaction of two single monomeric G-quadruplexes, but is a bimolecular inter-strand G4. This unique structural feature, along with the poor monomer–dimer interconversion, reasonably suggests that they might be derived from different folding pathways likely involving other unrevealed DNA intermediates. To test this hypothesis, we have observed the kinetics of the conformational changes of kit2 induced by the addition of a physiological concentration (150 mM) of KCl by CD spectroscopy (Figure 2).
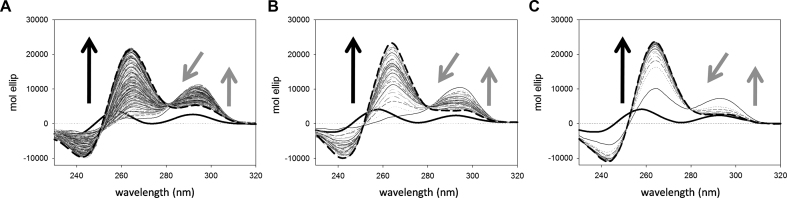
Figure 2.
Time dependent changes of CD spectra of 4 μM kit2 induced by the addition of 150 mM KCl in 10 mM Tris pH 7.5, at (A) 10°C, (B) 25°C and (C) 37°C. Data were collected for 27 h. Arrows indicate the variation of the main peaks as a function of time, solid and dashed lines correspond to the oligonucleotide in the absence of metal ion and at the end of the process, respectively.
In the absence of potassium ions, the CD spectrum of kit2 is not that expected for an unfolded sequence, since it exhibits two positive peaks at 258 and 298 nm (Figure 2, solid black lines). Immediately after the addition of the metal ion, a positive peak centered at 294 nm increased whereas the positive peak at 258 nm decreased. Subsequently, the contribution at 294 nm lowered and an intense positive band at 264 nm appeared, thus leading to the final dichroic profile of kit2. According to gel electrophoresis and AUC results, this final spectrum is comprised of contributions from both the monomeric and the dimeric forms (Figure 2, dashed line). Indeed, as previously reported, both of them exhibit a positive peak at 264 nm and a negative one at 245nm (22). As expected, by increasing the temperature, the processes became faster. Nevertheless, the overall spectral datasets (Figure 2) showed the same behavior, thus suggesting that the oligonucleotide undergoes the same structural variations at all the tested temperatures.
This first data set showed that the folding process of kit2 in KCl is not a one-step process, but that it is comprised of at least two processes: an initial fast folding into a kinetic intermediate that then converts into the thermodynamically stable structures. These two steps were then subsequently analysed separately.
The fast folding step analysis
As described above, the addition of KCl caused a rapid structural rearrangement of kit2 that occurred within the mixing time and it was not possible to characterize it by our CD equipment. In order to properly characterize this intermediate, stopped-flow experiments with UV detection were done. The dead time mixing of potassium was 10–20 ms, allowing for the resolution of faster steps. The absorbance variation in the 270–330 nm range was thus recorded over time.
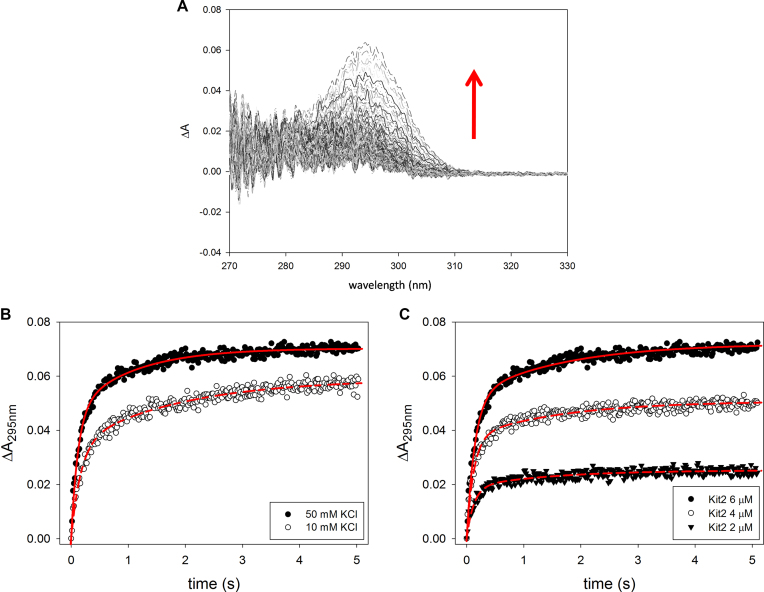
This approach allowed us to identify a major hyperchromic effect at 295 nm (Figure 3A) that is consistent with the formation of a G-quadruplex structure (27). Thus, in order to derive the kinetic parameters relative to the tetrahelix formation and to minimize the contribution of other forms to the data processing, we analysed the UV variation at 295 nm as a function of the incubation time. Additionally, the stopped-flow experiments were performed at 10 mM and 50 mM KCl to determine if this initial step required potassium (Figure 3B). All experimental data were analysed according to various kinetic models and, as supported by the Shapiro–Wilk normality test, the best fit was always obtained using a two-step exponential model (Equation 1, Figure 3B, red lines) (28). The derived kinetic constants are reported in Table 2. They indicate that within few seconds after the addition of K+, kit2 undergoes to at least two conformational changes (leading to forms thereafter named I1 and I2). Interestingly, with respect to G-quadruplex formation, the folding process is faster when the potassium concentration is increased, as reported for telomeric sequences (29,30).
Figure 3.
Time dependence variation of UV absorbance of kit2 upon addition of KCl in 10 mM Tris pH 7.5, 25°C. (A) Changes of UV spectra of 6 μM kit2 recorded after addition of 50 mM KCl (data were recorded for 5 s), (B) variation of the absorbance at 295nm induced by different concentrations of potassium ion (10 and 50 mM) and (C) using different concentrations of kit2 (2, 4, 6 μM) in 50 mM KCl.
Table 2. Kinetic constants derived by fitting the absorbance variation recorded at 295 nm after the addition of KCl using a ‘two kinetic processes’ model (Equation 1). Data were acquired in 10 mM Tris at 25°C.
| [KCl] | [kit2] | k 1 (s-1) | k 2 (s-1) |
|---|---|---|---|
| 10 mM | 6 μM | 6.00 ± 0.33 | 0.54 ± 0.04 |
| 50 mM | 6 μM | 6.96 ± 0.20 | 0.62 ± 0.03 |
| 50 mM | 4 μM | 8.13 ± 0.34 | 0.62 ± 0.04 |
| 50 mM | 2 μM | 8.12 ± 0.62 | 0.70± 0.09 |
To check if these initial folding steps were DNA concentration-dependent, stopped-flow experiments were repeated at 2, 4 and 6 μM of oligonucleotide. The results are reported in Figure 3C. As expected, by increasing the amount of DNA, the recorded UV signals increased whereas the derived kinetic constants did not change proportionally with the oligonucleotide concentration (Table 2). This is evidence that the species involved along these initial steps are monomeric.
The slow folding step analysis
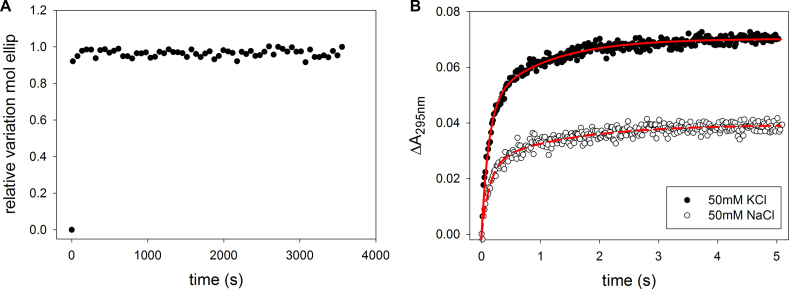
Since our evidence indicated that the very first part of the folding process is complete within 5 seconds which represent the lag-time for our CD experimental protocol, we were only able to use CD for a fine analysis of the slower step. Data were acquired in 50 mM KCl at different oligonucleotide concentrations (20 and 6 μM, Supplementary Figure S1). Immediately after the addition of potassium, the concentration of DNA in solution did not affect the molar ellipticity at 294 nm that reached a constant intensity of about 11000 deg·cm2·dmol−1. This data perfectly matches with our stopped-flow kinetic experiments performed at different oligonucleotide concentrations and further supports the hypothesis that in the first fast-steps no dimeric species are formed.
Interestingly, after complete equilibration (∼2.5 h), the molar ellipticity at 264 nm was much more intense in 20 μM kit2. This is not unexpected since the dimer should provide a higher dichroic signal than the monomer (31). In our system, this phenomenon was associated with a decrease of the signal at 294 nm that was directly related to the oligonucleotide concentration. Nevertheless, it remains much lower than the change at 264 nm. Thus, to analyse the slow kinetic rearrangements of our sequence, we took into account the signal at the lower wavelength. Considering the monomer–dimer formation, the experimental data points were well fitted by using an equation that contains first order (k4) and second order (k3) kinetic processes (Equation 2, see Materials and Methods). The derived constants are summarized in Table 3.
Table 3. Kinetic constants derived by applying a ‘first order + second order kinetic processes’ fitting model (Equation 2) to the dichroic signal variation at 264 nm of kit2 at different concentrations (6 μM and 20 μM) induced by the addition of 50 mM KCl in 10 mM Tris pH 7.5, at 25°C.
| [cKit2] | k 3 (s-1M-1) (·102) | k 4 (s-1) (·102) |
|---|---|---|
| 6 μM | 0.33 ± 0.07 | 0.040 ± 0.008 |
| 20 μM | 7.30 ± 1.86 | 0.076 ± 0.002 |
In both conditions, the first process (k3) is much faster than the second one. Interestingly this constant is strongly influenced by the amount of kit2 in solution (Table 3). This suggests that the process associated with k3 is the one responsible for the dimer formation.
Profiling the slower process using SVD analysis
Due to the simultaneous formation of monomeric and dimeric G4s in the slower steps, it was worthwhile to derive not only the kinetic parameters but also a reliable prediction of the number of participating species in solution as well as their basic dichroic spectra. Thus, we decided to apply Singular Value Decomposition (SVD) analysis to the data matrix formed by the time-dependent CD spectra of kit2 in the whole wavelength range. Indeed, SVD is an analytical tool that is very useful for characterizing complex kinetics events (24–26). In order to refer our analysis to kit2 behavior in a more physiological environment, we chose to perform SVD analysis on a dataset obtained in 150 mM KCl. Since to perform an accurate SVD analysis, we needed to acquire significant data points for all the forms involved in the folding process, we decided to work at 10°C. Indeed, as above reported (Figure 2), by reducing the temperature the folding process becomes slower but the conformational changes are conserved.
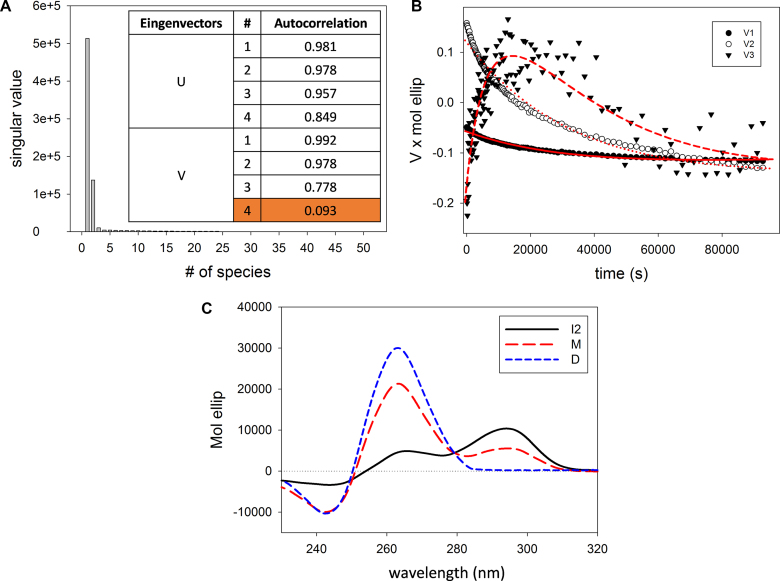
In agreement with the result of data analysis performed at a single wavelength, the singular values contained in the S matrix as well as the U and V autocorrelation coefficients (meaningful above 0.75) indicated that there are at least three species in solution that contribute to the variation of the CD signal (Figure 4A). These species should account for the thermodynamically stable structures (the monomeric and the dimeric G-quadruplexes observed in AUC and PAGE) as well as for the initial intermediate, derived from the fast-initial steps. The significant V eingenvectors were well fitted by Equation (2) (Figure 4B). This model provided the kinetic constants (Table 4) and the parameters forming the H matrix. The linear combination of H matrix and U × S matrix provides the actual spectra of the species recurring along the process.
Figure 4.
(A) S matrix values and autocorrelation coefficients of U and V matrices indicating the relevance of species in solution participating to the overall dichroic signal variations, (B) plot of the significant V eingenvectors as a function of time and their global fitting (red line) (Equation 2, see Materials and Methods), (C) CD basic spectra of the species in solution that contribute to the overall changes of the dichroic signal as derived from SVD analysis, where I2 refers to the second intermediate, M to the final monomeric species and D to the dimer.
Table 4. Fitting parameters derived by applying a ‘first order + second order independent processes’ kinetic model (Equation 2) to the significant V-eingenvectors obtained using SVD analysis. Spectra dataset was acquired upon the addition of 150 mM KCl in 10 mM Tris at 10°C.
| Fitting model | R 2 | Kinetic constants (·105) |
|---|---|---|
| First order + Second order independent processes | 0.91 | k 3 = 9.02 ± 3.52 s-1 M-1 |
| k 4 = 2.90 ± 0.20 s-1 |
The so obtained spectral shapes are reported in Figure 4C. One species presents a positive peak at 294 nm and a shoulder at 270 nm. It corresponds to the starting point of these kinetic experiments and we can relate it to the second folding intermediate formed at the end of the fast folding phase (I2). Its dichroic features indicate it as a hybrid G-quadruplex.
The other two derived spectra are attributable to the thermodynamically stable species, the monomer and the dimer. Both present a main positive chiroptical contribution at 264 nm and a minor negative one at 245 nm, in line with their parallel conformation. As reported in the literature, due to the higher number of bases participating in the structure formation, the dimeric species is expected to exhibit a more intense dichroic signal (31). This observation confirms that the highest dichroic signal is related to the dimeric form.
Characterization of the species in sodium-containing solution
It is well documented that different ionic species can contribute to G-quadruplex formation while often leading to different specific structures. Even though sodium is less representative of the physiological environment in the nucleolus, this cation can interfere in the final folding of the studied sequence. Thus, we evaluated the ability of sodium ions to induce kit2 to assume a G-quadruplex arrangement.
Electrophoretic resolution showed that when kit2 was annealed in the presence of sodium, it formed one fast electrophoretic band corresponding to the monomeric G4 characterized in K+ but no multimeric species (Figure 5A). Analytical ultracentrifugation results were in agreement: they confirmed that NaCl changes the hydrodynamic volume of the oligonucleotide and leads to a distribution of the sedimentation coefficients compatible with the monomeric oligonucleotide (Figure 5B). Nevertheless, the shape of the curve appeared to broaden, thus suggesting the presence of a mixed population that includes more than one monomeric form of kit2.
Figure 5.
(A) Electrophoretic resolution of kit2 annealed under different ionic conditions (no cations, 150 mM KCl, 150 mM NaCl) performed on 15% acrylamide native PAGE in 1× TBE using a scrambled oligonucleotide of the same molecular weight as marker (M), (B) relative distribution of kit2 species in solution as function of their sedimentation coefficient with/without 50 mM NaCl (respectively, dashed and solid lines) in 10 mM Tris pH 7.5.
In order to determine if this profile is comprised of a mixture of more than one G4 form or of unfolded/folded species, the CD spectrum of kit2 was recorded in the presence of increasing concentration of sodium chloride until a plateau was reached.
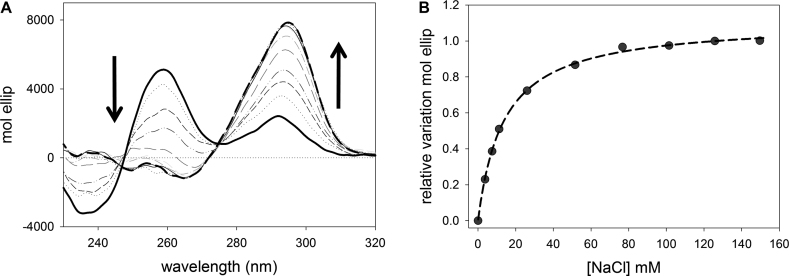
As reported in Figure 6A, the CD spectrum of the pre-folded kit2 changed upon NaCl addition, and showed the formation of a positive band around 294 nm and a negative band at 264 nm. These are the expected features of an antiparallel G-quadruplex. During CD titrations, an isodichroic point was maintained at 275 nm. Accordingly, the relative variation of the signal recorded at 294 nm as function of the NaCl (Figure 6B) was well fitted using a ‘one binding site model’. The derived KDapp value was 13.69 ± 0.47 mM, higher than the value reported for KCl (9.49 ± 0.83 mM) (22). Also the intensity of the final dichroic signal is lower in Na+ than in K+, thus suggesting that in sodium the G-quadruplex form of kit2 is less rigidly organized. This data is consistent with a general lower efficiency of Na+ versus K+ in the stabilization of G-quadruplex structures. When this observation is coupled with the evidence that in 50 mM NaCl kit2 is not completely folded into a G4 (approximately the 15% maintains the pre-folded form), it becomes easier to justify the low resolution of monomeric components derived by AUC.
Figure 6.
(A) CD spectra representing the titration of 4 μM kit2 with increasing concentration of NaCl (0–150 mM) in 10 mM Tris pH 7.5 at 25°C and (B) relative variation of molar ellipticity obtained monitoring the spectral changes induced by the addition of NaCl at 294 nm.
Folding kinetics of kit2 in sodium
In order to assess how the presence of Na+ also impacts the folding kinetics of kit2, a comparable spectroscopic approach (CD kinetics and stopped-flow UV analyses) was performed.
When we explored the fast processes by stopped-flow UV, we observed that the addition of NaCl induced effects comparable to KCl (Figure 7B). Indeed, also in this case a ‘two kinetic processes model’ (Equation (1)) properly fitted the absorbance variation at 295 nm as a function of time (Figure 7B). The kinetic parameters are summarized in Table 5.
Figure 7.
(A) Kinetic profile of kit2 folding after the addition of NaCl (final concentration 50 mM) monitoring the variation of molar ellipticity at 294 nm, (B) absorbance variation at 295 nm of 6 μM kit2 induced by the addition of different metal ions (50 mM sodium or potassium chloride) plotted over time recorded in 10 mM Tris, pH 7.5, 25°C.
Table 5. Kinetic constants derivedby applying a ‘two kinetic processes’ (Equation 1) fitting model to the absorbance variation recorded at 295 nm for kit2 (6 μM) upon the addition of 50 mM NaCl, in 10 mM Tris, pH 7.5, 25°C.
| [NaCl] | k 1’ (s-1) | k 2’ (s-1) |
|---|---|---|
| 50 mM | 7.04 ± 0.49 | 0.71 ± 0.05 |
However, distinct from results in potassium-containing solution, in the presence of sodium no additional slow folding steps were detected, thus making the folding process very fast and fully completed in the timescale of few seconds (Figure 7A). This means that in sodium a reduced number of folding intermediates (named I1’ and I2’) is required to reach the final antiparallel G-quadruplex.
Sodium–potassium folding competition
The final folded state of kit2 in NaCl consists of an antiparallel G-quadruplex that does not correspond to other species identified along the potassium driven folding pathway. The physiological environment contains a complex mixture of different ions, and even though cells present a smaller amount of Na+ compared to the concentration of K+, the first cation can affect the overall arrangement of the oligonucleotide as well as the intermediates required to interconvert the sodium vs the potassium folded forms. Therefore, after the characterization of the oligonucleotide conformations in each cation species, we considered the folding of kit2 in solutions containing both the metal ions. We started from a solution of kit2 equilibrated in 50 mM sodium chloride to which we added an equimolar KCl concentration and the changes of the dichroic spectrum were recorded over time.
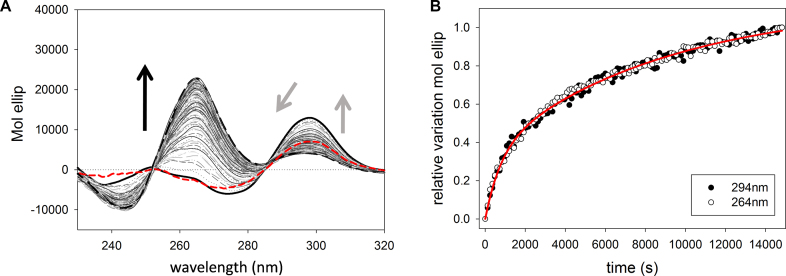
Upon addition of KCl, the starting spectrum corresponding to the Na+-stabilized antiparallel G-quadruplex (Figure 8A, red line), immediately changed (the timescale is comparable to the mixing time) triggering an increase of the signal at 294 nm and a decrease of the band at 275 nm. This fast ion exchange has already been observed on the telomeric sequence (32). When compared to the fast-forming intermediate characterized in KCl-containing solution, the main difference rests in the lack of any contribution at 260 nm. Its presence suggests that in these experimental conditions potassium can transiently favor or stabilize an antiparallel arrangement of kit2 (named I3), not previously detected along the KCl folding pathway in the absence of Na+. Also this intermediate then slowly interconverts to provide the expected spectrum for potassium containing solutions (a positive peak at 264 nm and a negative one at 245 nm). Again, this slower kinetic process was well described by a ‘first order + second order independent processes’ (Equation 2).
Figure 8.
(A) CD spectra acquired during the kinetic structural rearrangement of 6 μM kit2 after the addition of KCl to a final concentration of 50 mM in 10 mM Tris pH 7.5, 50 mM NaCl at 25°C (data were collected for 4 h). Dashed red line, solid black line and the dashed black line represent,respectively, kit2 folded in 50 mM in NaCl, immediately after the addition of 50 mM KCl and at the thermodynamic equilibrium. The corresponding relative variations of the signals recorded at 294 nm and at 264 nm are reported in panel (B).
Thus, the presence of sodium is not sufficient to impair the formation of the K+-induced parallel forms although it modulates the conformational equilibria of the sequence in solution. Indeed, as reported in Table 6, the rates of the slower steps are slower in Na+/K+ than in K+ alone.
Table 6. Kinetic constants provided by a ‘first order + second order independent processes’ fitting model (Equation 2) applied to the variation of molar ellipticity recorded over time at 264 nm and 294 nm of kit2 in 50 mM NaCl and 10 mM Tris upon the addition of 50 mM KCl at 25°C.
| Salt conditions | k 3’ (s-1M-1) (·102) | k 4’ (s-1) (·102) |
|---|---|---|
| 50 mM NaCl-50 mM KCl | 0.13 ± 0.01 | 0.011 ± 0.001 |
DISCUSSION
A key challenge in G4 ligand design concerns the polymorphic behavior of the selected targets. In fact, whereas high resolution techniques, like as NMR and X-ray are extensively applied to provide structural data related to the thermodynamically stable forms, information about G-quadruplex folding pathways are currently limited to a few examples (33). The folding of G4 structures in general is recognized to be kinetically complex and might be described by a number of theoretical concepts, including ‘folding funnels’, ‘kinetic partitioning’ or more conventional sequential or parallel kinetic reaction mechanisms (5,33–35). These are difficult to test and distinguish experimentally, especially because of the differing timescales accessible to different experimental or computational methods. What is clear is that folding of G4 structures is not a simple two-state process and that a complex array of intermediate states are populated along the folding pathway. These transient intermediates may play physiologically important roles and should not be neglected.
In this context, the results herein provide deep insights on kit2 folding equilibria.
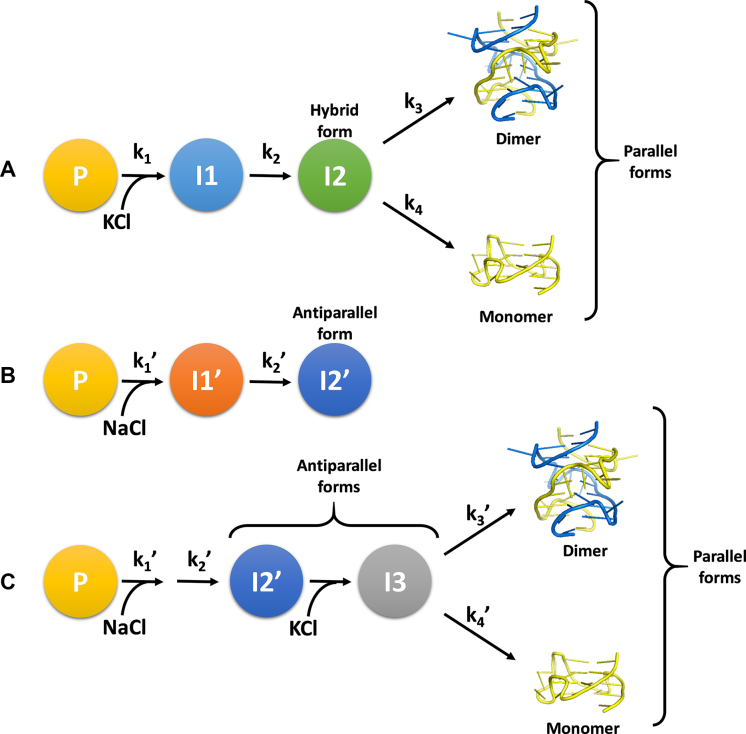
The cation-induced folding pathways of kit2 were shown to be very complex and greatly affected by the environmental conditions. A schematic illustration of the folding processes based on data presented herein is shown in Figure 9.
Figure 9.
(A) Potassium-induced folding pathway of kit2, (B) sodium-induced folding pathway, (C) sodium–potassium mixture folding pathway.
The first observation is that in the absence of cations, the sequence appears to be arranged in a pre-folded form (P) with spectroscopic and electrophoretic features completely different from those expected for an unfolded oligonucleotide. This behavior has been reported earlier for other G-rich sequences (29,36). We inferred that our experimental starting conditions (10 mM Tris) present a sufficient ionic strength to support the collapse of the unfolded oligonucleotide into a compact structure, e.g. hairpin (35,37,38). Upon the addition of cations to the solution, the electrostatic repulsion among the phosphate groups is strongly and quickly reduced. In accordance with electrolyte theory, this favors the formation of more structured species ultimately leading to the final G4 forms (39).
At the very early stage of cation-driven structural rearrangement of kit2, two fast conformational changes are detected either in the presence of sodium or potassium ions. UV stopped-flow measurements confirmed that these initial folding steps affect the absorbance signal at 295 nm and the derived kinetic constants are cation-concentration dependent. As far it concerns I1 and I1’ (the first intermediates in K+ and Na+, respectively) we did not succeed in obtaining structural information, but these spectral features are compatible with G-quadruplex-like structures. They could represent intermediates with less than a full stoichiometric complement of cations as already reported in the literature for the initial folding step of other G-rich sequences or triplex intermediates (29).
The formation constants for these initial species are only slightly different when measured in Na+ or K+. The same occurs for I2 and I2’, the first intermediates for which we succeeded in acquiring structural information. Under both experimental conditions, they derive from the full conversion of I1 or I1’, respectively. This is supported by their half-life timescale and, limited to K+ condition, by SVD analysis that excluded a significant presence of I1 in the slower part of the process. Despite these extensive similarities, I2 and I2’ do not belong to overlapping folding pathways. Indeed, in sodium, I2’ represents the thermodynamically stable structure and our data indicate it is a monomeric antiparallel G-quadruplex. This is in keeping with the behavior previously reported for the telomeric sequence (29). Conversely, the potassium-induced I2 is a short-lived intermediate corresponding to a hybrid G-quadruplex that slowly rearranges into the final forms.
Following the evolution of I2 on a time-scale of hours, SVD analysis showed that the kit2 energetic landscape in potassium does not favor the stability of the hybrid form. Indeed, it converts into the final parallel monomeric and dimeric forms stably trapped into two energetic minima. According to the data obtained by analytical ultracentrifugation, these two species correspond to the structures already reported in literature (21).
The plasticity of kit2 is further reinforced by exploring more complex ionic environments, where additional conformational rearrangements can be detected. Indeed, starting from the sodium-stabilized antiparallel G4 (I2’), the addition of potassium cations drives the process toward the formation of the same thermodynamically stable parallel G-quadruplexes. Interestingly, this process involves an unprecedented intermediate (I3), when compared to K+-containing conditions. It fully retains the antiparallel arrangement typically observed in Na+. This means that the oligonucleotide can follow multiple pathways to reach the final monomeric and the dimeric parallel structures.
As already reported for other sequences such as the telomeric one, we can propose that KCl efficiently replaces within the mixing time the sodium cations preserving the G-quadruplex core (40). Herein, the observed stabilization of the antiparallel form in potassium, underlined by the rapid increase of its dichroic signal after its addition, is the experimental evidence that this replacement occurs. Subsequently, this unique potassium-stabilized antiparallel G-quadruplex (I3) acts as short-lived intermediate that directly leads to the formation of the final monomer and dimer. Thus, in this experimental condition, the previous identified K+-induced intermediate I2 is not involved in the oligonucleotide folding process.
It is worthwhile to compare our model to those derived for the widely studied telomeric sequence (5,33). A branched pathway is required to describe our data. A major difference is that we were not able to identify antiparallel intermediates in pure K+ solutions. Nevertheless, we cannot exclude the involvement of this species since it appears to represent the main component in the Na+/K+ conformational switch (40). Such an highly polymorphic behavior for kit2 has been proposed to be enabled by the long and unstructured central loop that allows it to adopt either an all-parallel, propeller type conformation, or a mixed parallel/antiparallel conformation (20).
In conclusion, based on the data presented herein, it appears that the wild-type kit2 sequence also converges toward two shared main G-quadruplex folded forms. It emerges that the population of G-quadruplex or G-quadruplex-like conformations in solution is comprised of multiple components and that its composition significantly varies with time and environmental conditions. The presence of several relevant folding intermediates, some of them characterized by a half-life within the timescale of physiological DNA processing events, definitely supports their potential role in gene regulatory processes. Thus, taking into account only the high-resolution structures of kit2 as the actual G4 elements working during the transcription, leads to a loss of information regarding the functional arrangement of the gene promoter.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Del Villar-Guerra R. for support with data analysis.
Footnotes
Present address: Claudia Sissi, Department of Pharmaceutical and Pharmacological Sciences, University of Padova, 35131 Padova, Italy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Padova [CPDA147272 to C.S., PhD fellowships to R.R.]; National Institutes of Health [GM077422 to J.B.C.]. Funding for open access charge: CS personal research funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004; 43:668–698. [DOI] [PubMed] [Google Scholar]
- 3. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug. Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell. Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 5. Gray R.D., Trent J.O., Chaires J.B.. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014; 426:1629–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maiuri P., Knezevich A., De Marco A., Mazza D., Kula A., McNally J.G., Marcello A.. Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 2011; 12:1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ben-Ari Y., Brody Y., Kinor N., Mor A., Tsukamoto T., Spector D.L., Singer R.H., Shav-Tal Y.. The life of an mRNA in space and time. J. Cell. Sci. 2010; 123:1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morisaki T., Muller W.G., Golob N., Mazza D., McNally J.G.. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014; 5:4456–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karpova T.S., Kim M.J., Spriet C., Nalley K., Stasevich T.J., Kherrouche Z., Heliot L., McNally J.G.. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008; 319:466–469. [DOI] [PubMed] [Google Scholar]
- 10. Sharp Z.D., Mancini M.G., Hinojos C.A., Dai F., Berno V., Szafran A.T., Smith K.P., Lele T.P., Ingber D.E., Mancini M.A.. Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J. Cell. Sci. 2006; 119:4101–4116. [DOI] [PubMed] [Google Scholar]
- 11. Bosisio D., Marazzi I., Agresti A., Shimizu N., Bianchi M.E., Natoli G.. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 2006; 25:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McNally J.G., Muller W.G., Walker D., Wolford R., Hager G.L.. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000; 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- 13. Gebhardt J.C., Suter D.M., Roy R., Zhao Z.W., Chapman A.R., Basu S., Maniatis T., Xie X.S.. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods. 2013; 10:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Metcalfe D.D. Mast cells and mastocytosis. Blood. 2008; 112:946–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gregory-Bryson E., Bartlett E., Kiupel M., Hayes S., Yuzbasiyan-Gurkan V.. Canine and human gastrointestinal stromal tumors display similar mutations in c-KIT exon 11. BMC Cancer. 2010; 10:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zorzan E., Da Ros S., Musetti C., Shahidian L.Z., Coelho N.F., Bonsembiante F., Letard S., Gelain M.E., Palumbo M., Dubreuil P. et al. . Screening of candidate G-quadruplex ligands for the human c-KIT promotorial region and their effects in multiple in-vitro models. Oncotarget. 2016; 7:21658–21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLuckie K.I., Waller Z.A., Sanders D.A., Alves D., Rodriguez R., Dash J., McKenzie G.J., Venkitaraman A.R., Balasubramanian S.. G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J. Am. Chem. Soc. 2011; 133:2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gunaratnam M., Swank S., Haider S.M., Galesa K., Reszka A.P., Beltran M., Cuenca F., Fletcher J.A., Neidle S.. Targeting human gastrointestinal stromal tumor cells with a quadruplex-binding small molecule. J. Med. Chem. 2009; 52:3774–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller M.C., Le H.T., Dean W.L., Holt P.A., Chaires J.B., Trent J.O.. Polymorphism and resolution of oncogene promoter quadruplex-forming sequences. Org. Biomol. Chem. 2011; 9:7633–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu S.T., Varnai P., Bugaut A., Reszka A.P., Neidle S., Balasubramanian S.. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009; 131:13399–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuryavyi V., Phan A.T., Patel D.J.. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010; 38:6757–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Da Ros S., Zorzan E., Giantin M., Zorro Shahidian L., Palumbo M., Dacasto M., Sissi C.. Sequencing and G-quadruplex folding of the canine proto-oncogene KIT promoter region: might dog be used as a model for human disease?. PLoS One. 2014; 9:e103876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Greenfield N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2006; 1:2891–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendler R.W., Shrager R.I.. Deconvolutions based on singular value decomposition and the pseudoinverse: a guide for beginners. J. Biochem. Biophys. Methods. 1994; 28:1–33. [DOI] [PubMed] [Google Scholar]
- 25. Gray R.D., Chaires J.B.. Analysis of multidimensional G-quadruplex melting curves. Curr. Protoc. Nucleic Acid Chem. 2011; doi:10.1002/0471142700.nc1704s45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DeSa R.J., Matheson I.B.. A practical approach to interpretation of singular value decomposition results. Methods Enzymol. 2004; 384:1–8. [DOI] [PubMed] [Google Scholar]
- 27. Mergny J.L., Li J., Lacroix L., Amrane S., Chaires J.B.. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005; 33:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peat J., Barton B.. Medical Statistics: A Guide to Data Analysis and Critical Appraisal. 2008; 1st edn, Blackwell Publishing. [Google Scholar]
- 29. Gray R.D., Chaires J.B.. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008; 36:4191–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ying L., Green J.J., Li H., Klenerman D., Balasubramanian S.. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14629–14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tothova P., Krafcikova P., Viglasky V.. Formation of highly ordered multimers in G-quadruplexes. Biochemistry. 2014; 53:7013–7027. [DOI] [PubMed] [Google Scholar]
- 32. Gray R.D., Petraccone L., Trent J.O., Chaires J.B.. Characterization of a K+-induced conformational switch in a human telomeric DNA oligonucleotide using 2-aminopurine fluorescence. Biochemistry. 2010; 49:179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marchand A., Gabelica V.. Folding and misfolding pathways of G-quadruplex DNA. Nucleic Acids Res. 2016; 44:10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bessi I., Jonker H.R., Richter C., Schwalbe H.. Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem. Int. Ed. Engl. 2015; 54:8444–8448. [DOI] [PubMed] [Google Scholar]
- 35. Stadlbauer P., Kuhrova P., Banas P., Koca J., Bussi G., Trantirek L., Otyepka M., Sponer J.. Hairpins participating in folding of human telomeric sequence quadruplexes studied by standard and T-REMD simulations. Nucleic Acids Res. 2015; 43:9626–9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ceru S., Sket P., Prislan I., Lah J., Plavec J.. A new pathway of DNA G-quadruplex formation. Angew. Chem. Int. Ed. Engl. 2014; 53:4881–4884. [DOI] [PubMed] [Google Scholar]
- 37. Murthy V.L., Rose G.D.. Is counterion delocalization responsible for collapse in RNA folding?. Biochemistry. 2000; 39:14365–14370. [DOI] [PubMed] [Google Scholar]
- 38. Thirumalai D., Hyeon C.. RNA and protein folding: common themes and variations. Biochemistry. 2005; 44:4957–4970. [DOI] [PubMed] [Google Scholar]
- 39. Manning G.S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 1978; 11:179–246. [DOI] [PubMed] [Google Scholar]
- 40. Gray R.D., Li J., Chaires J.B.. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009; 113:2676–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.