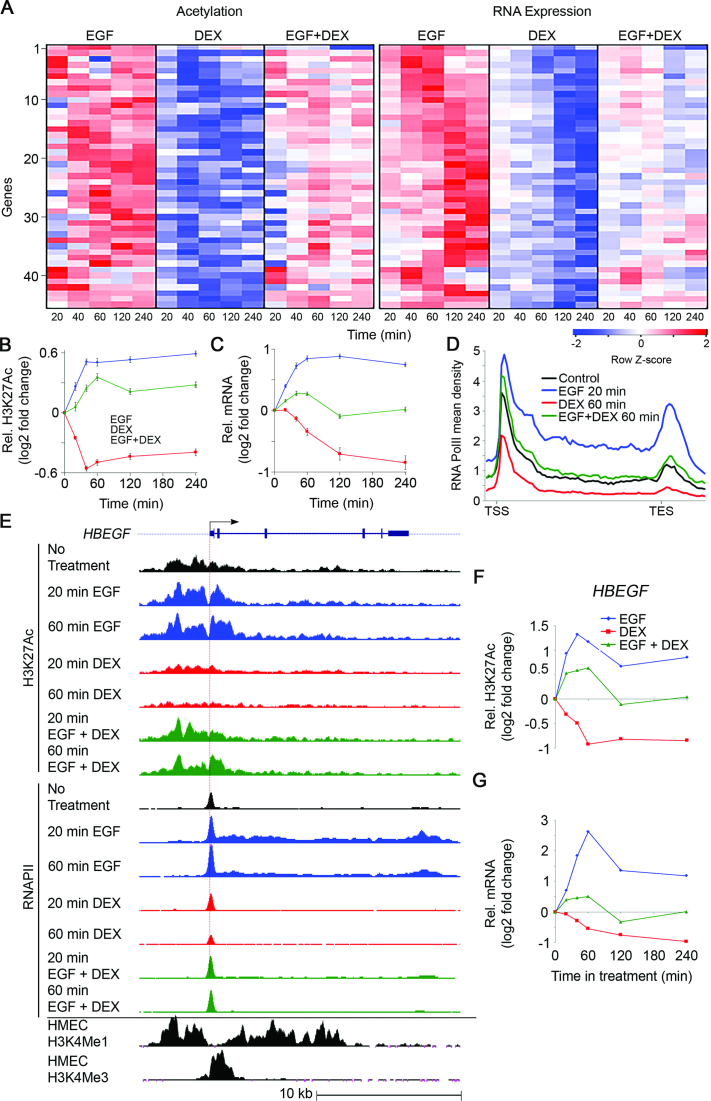
Figure 1.
The group of genes whose mRNAs are upregulated by EGF and downregulated by DEX (module B) displays concordant dynamic alterations of transcript abundance, H3K27Ac and RNA polymerase II occupancy. Serum-starved MCF10A cells were treated with EGF (10 ng/ml), DEX (100 nM) and the combination, for the indicated time intervals. (A) A heatmap showing alterations in H3K27Ac occupancy at promoter regions and RNA abundance of 45 Module B genes. (B and C) Time series showing average alterations in H3K27Ac occupancy (B) and mRNA abundance (C) of the 45 module B genes. (D) Shown are average profiles of RNAPII ChIP-Seq across all module B genes. Mean density was calculated as the average read number normalized to sequencing depth (total bin number: 85, from −1 kb upstream to the TSS to +3 kb after the TES). RNAPII profiles represent the average of biological duplicates. (E) The genomic structure and TSS of HBEGF are shown (top). Also shown are H3K27Ac and RNAPII tracks representing the normalized ChIP-seq read coverage. The lowermost two tracks show published ChIP-seq data of H3K4Me1 and H3K4Me3 for human mammary epithelial cells (wgEncodeEH000089 and wgEncodeEH000091, respectively). (F and G) Time series showing alterations in H3K27Ac occupancy (F) and transcript abundance (G) of the HBEGF gene. Bars represent standard errors of the mean of fold change.