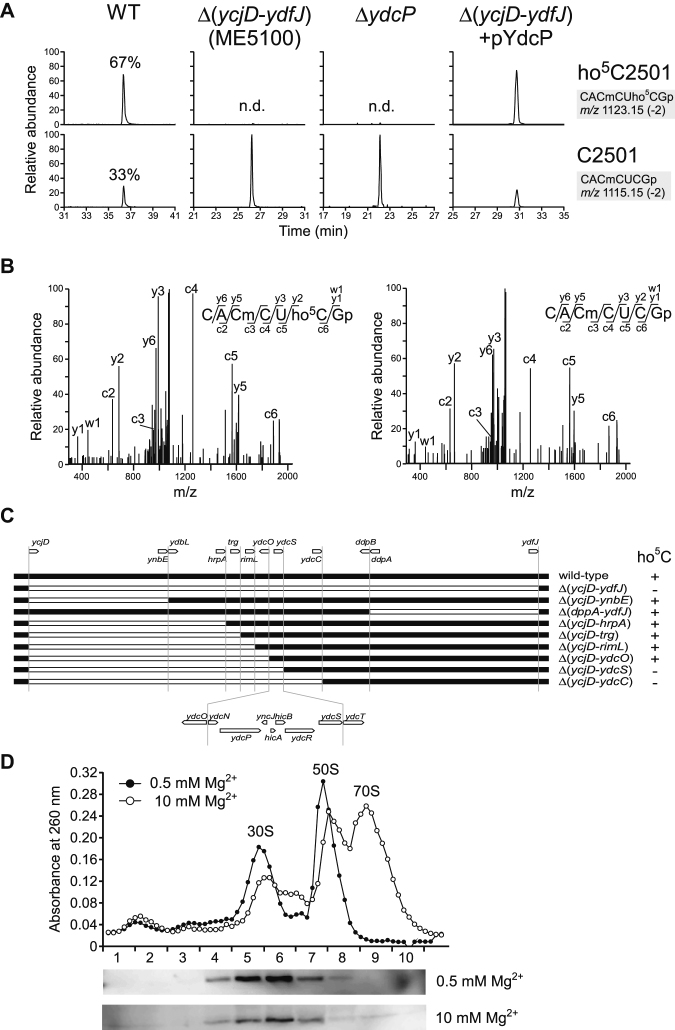
Figure 2.
RlhA/YdcP is responsible for ho5C2501 formation. (A) RNA-MS of 48-mer segments of 23S rRNA from WT (leftmost panels), ME5100 (left panels), ΔydcP (right panels) and ME5100 rescued by plasmid-encoded ydcP (rightmost panels). Shown are extracted mass chromatograms (XICs) for divalent negative ions of the RNase T1–digested heptamer fragments with (upper panels) or without (lower panels) ho5C2501. Frequencies of ho5C2501 are indicated. n.d., not detected. (B) Collision-induced dissociation (CID) spectra of the heptamer fragments with (left panel) or without (right panel) ho5C2501. A divalent ion for each fragment (m/z 1123.1 and 1115.1) was employed as the parent ion for CID. c, y, and w series product ions are assigned in each sequence. (C) E. coli genomic region from ycjD to ydfJ, and a series of deletion strains used to narrow down the gene responsible for ho5C2501 formation. The white bar represents a deleted region. The presence (+) or absence (–) of ho5C2501 is denoted in each strain. (D) Sucrose density gradient profiling of SPA-tagged RlhA in ribosomal fractions. Upper panel shows UV traces at 260 nm for the ribosomal fractions in 0.5 (filled circles) and 10 mM (open circles) Mg2+ conditions. Lower panels show western blotting to detect SPA-tagged RlhA protein in each Mg2+ concentration.