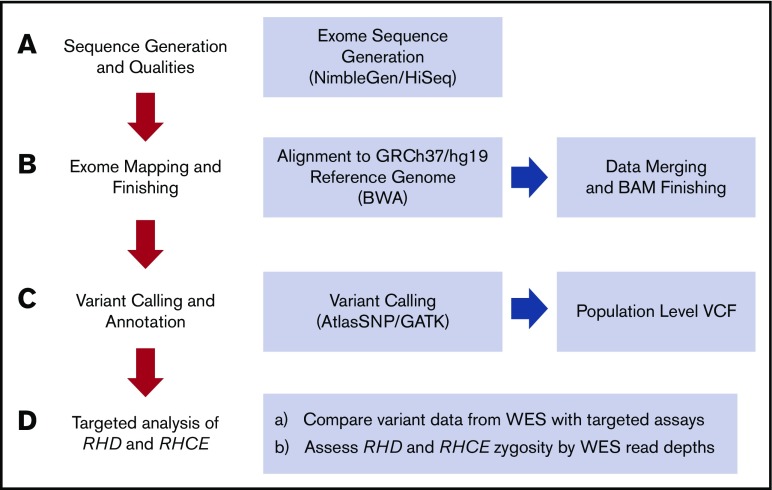
Figure 2.
Mercury analysis pipeline. (A) Raw data from the sequencing instrument is passed to primary analysis software to generate sequence reads and base call confidence values (qualities). (B) Reads and qualities are passed along to a mapping tool Burrows-Wheeler algorithm (BWA) for comparison with a reference genome. The placement of reads on the reference genome produces a Binary-format Sequence Alignment Map (BAM) file and individual event BAMs were merged to make a single sample-level BAM file. (C) AtlasSNP and Genome Analysis Toolkit (GATK) are used to identify variants and produce annotated variant call files (VCFs). (D) In this study, we specifically interrogated WES data for the RHD and RHCE genes compared with conventional targeted assays.