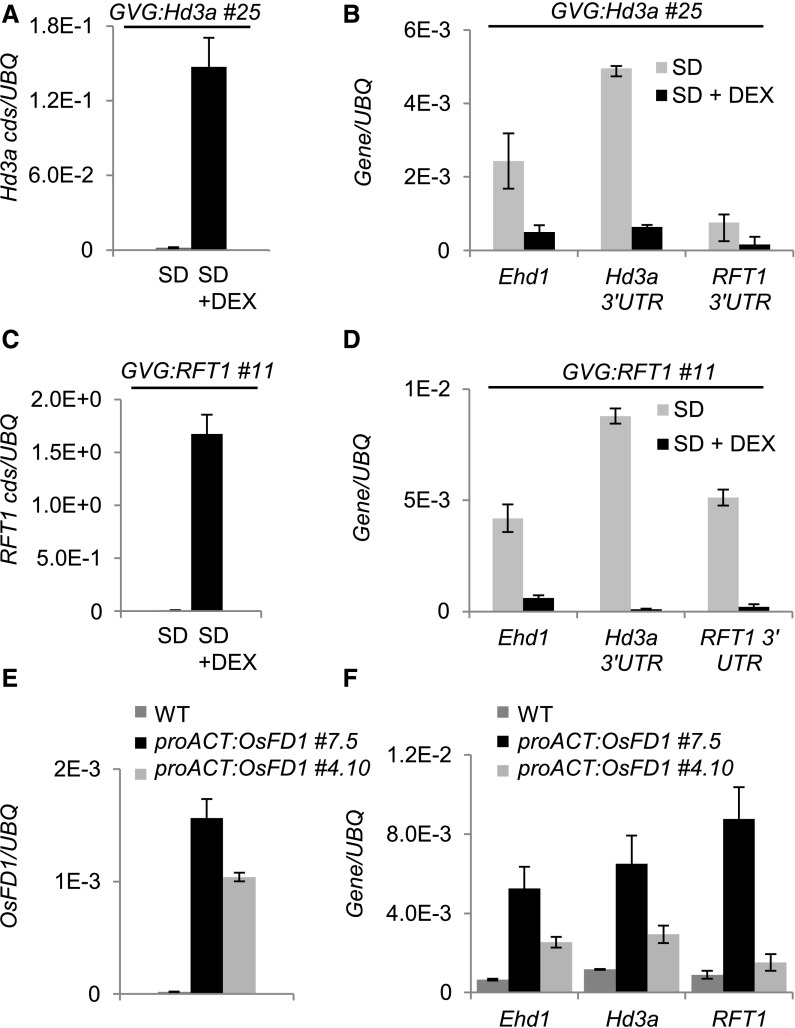
Figure 3.
A Negative Feedback Loop Independent of OsFD1 Reduces Ehd1, Hd3a, and RFT1 Expression during Floral Induction in Leaves.
(A) to (D) DEX-induced overexpression of Hd3a ([A] and [B]) or RFT1 ([C] and [D]) causes strong increase of Hd3a (A) or RFT1 (C) transcript accumulation from transgenic sequences, but downregulation of Ehd1, Hd3a, and RFT1 endogenous transcripts, compared with mock-treated controls ([B] and [D]).
(E) and (F) Two independent transgenic proACT:OsFD1 lines show increased expression of OsFD1 (E) and of Ehd1, Hd3a, and RFT1 in leaves compared with the wild type (F). DEX was applied at 13 DAS, and leaf samples were collected at ZT0, 16 h later. proACT:OsFD1 plants were collected at ZT0 and 12 DAS. Leaves from 10 plants per treatment were sampled. UBQ was used as standard for quantification of gene expression. Data are represented as mean ± sd. Primers on Hd3a or RFT1 coding sequences or on the 3′ untranslated regions were used to distinguish transgenic+endogenous ([A] and [C]) from endogenous transcripts, respectively ([B] and [D]). ANOVA tests for graphs in (A) to (F) are shown in Supplemental File 1.