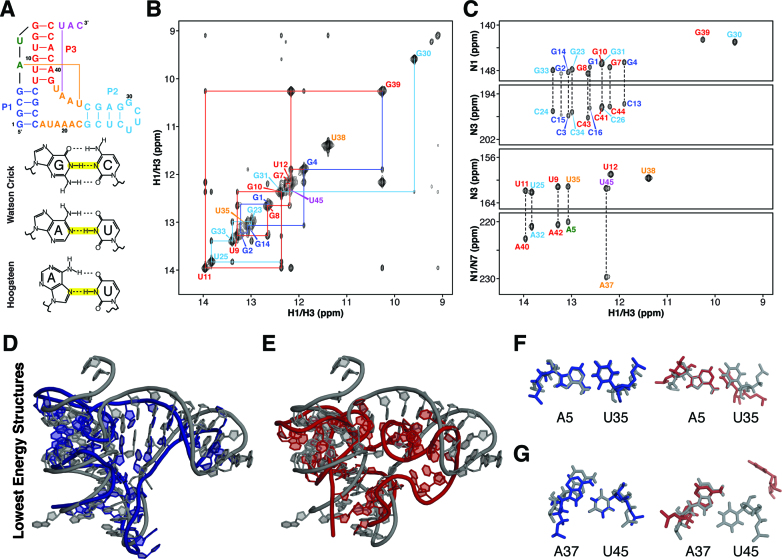
Figure 3.
Refinement of B. cereus fluoride riboswitch aptamer based on NMR imino-NOE constraints. (A) Secondary structure of a Bacillus cereus fluoride riboswitch. (B) NMR 1H–1H NOESY spectrum of fluoride-bound B. cereus riboswitch. (C) NMR JNN-COSY spectrum of fluoride-bound B. cereus riboswitch. (D) Crystal structure of the T. petrophila fluoride riboswitch (PDB ID: 4ENC) in gray aligned with lowest energy iFoldNMR model of B. Cereus fluoride riboswitch aptamer. (E) Crystal structure aligned with structural model constrained by secondary structure information only. (F) A5-U35 reverse Watson–Crick base pair. (G) A37-U45 reverse Hoogsteen base pair.