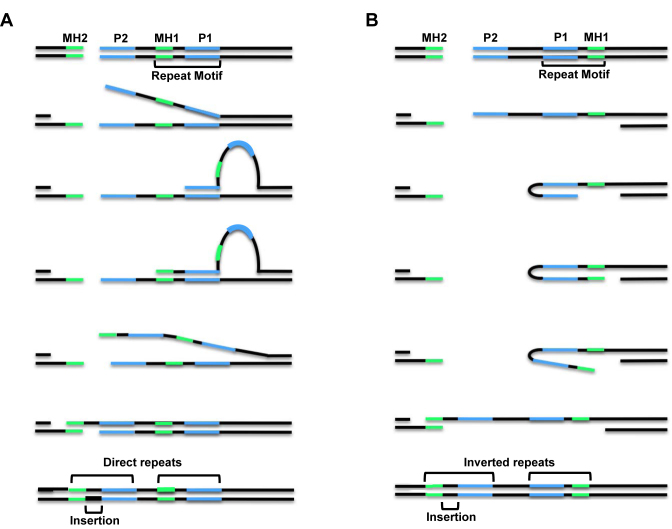
Figure 1.
The SD-MMEJ model for alternative end joining repair. (A) Loop-out mechanism with DNA unwinding prior to loop formation. (B) Snap-back mechanism with DNA resection prior to hairpin formation. Both mechanisms utilize annealing of break-proximal primer repeats (P2) to break-distal primer repeats (P1), which primes nascent synthesis that can lead to insertions (black) and the creation of new microhomologous sequences (MH1, green). For loop-out SD-MMEJ, P1 and P2 are direct repeats, while for snap-back SD-MMEJ they are inverted repeats. Repair concludes with unwinding of secondary structures, annealing of nascent microhomologies with MH2 sequences on the other side of the break, fill-in synthesis, and ligation. For the repair events shown here, the inserted sequence becomes part of new, longer direct or inverted repeats. Not shown are the trimming of non-homologous flap intermediates when P2 and MH2 are not directly adjacent to the break site, or deletion junctions with no net insertion that are formed when P1 and MH1 are directly adjacent to each other.