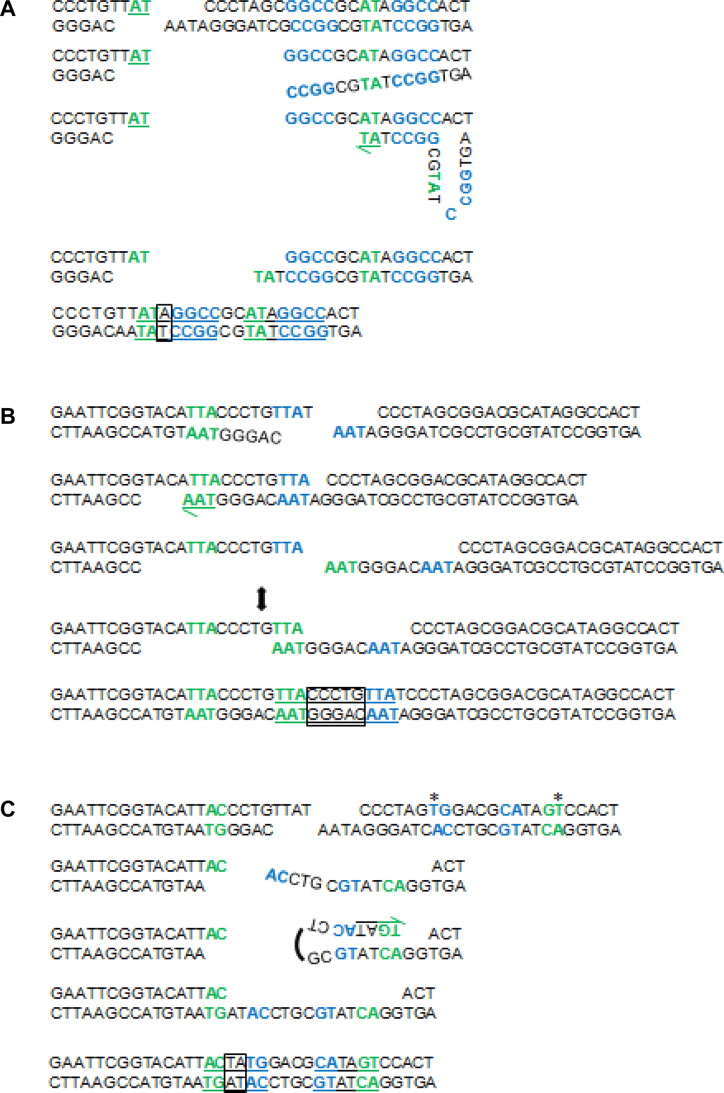
Figure 9.
Models for SD-MMEJ consistent indel junctions. Shown are primer repeats (blue), microhomology repeats (green), insertions (boxes), and final repeat motif (underlined). (A) Iw7 loop-out event. Unwinding/resection of DNA on the right side of the break allows primer repeats to anneal, followed by limited synthesis, dissociation, and annealing of microhomology repeats. Endonuclease/flap cleavage events are not shown for simplicity. (B) M1 trans SD-MMEJ event. The two-ended arrow indicates transition of the TTA primer repeat to a microhomology repeat. This junction can also be explained by a snap-back mechanism on the left side of the break. (C) M2 snap back event. Asterisks indicate engineered sequence changes utilized during repair. Note that 3′→5′ resection or endonuclease activity following the annealing of primer repeats is required prior to initial synthesis.