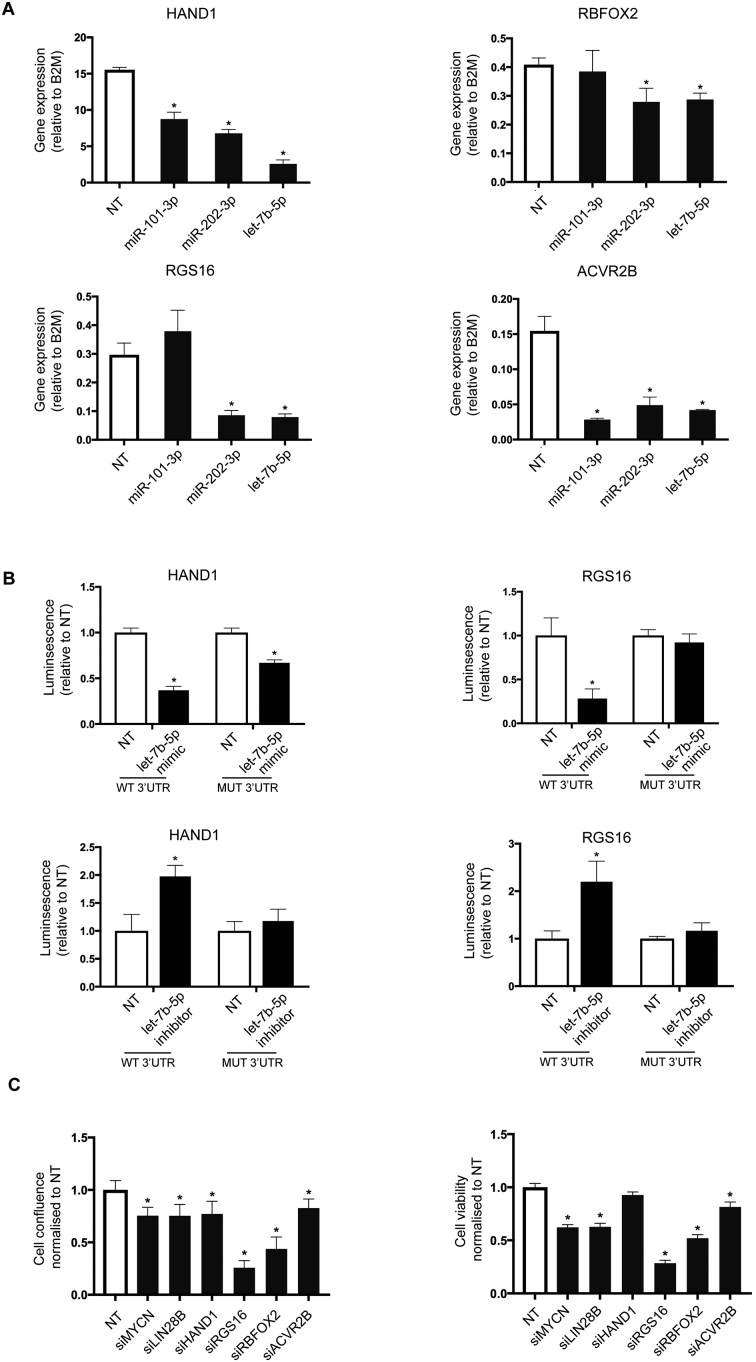
Figure 5.
Novel let-7 targets regulate the survival of neuroblastoma cancer cells. (A) KELLY cells were transfected with let-7b-5p, miR-101-3, miR-202-3p or NT, and changes in mRNA levels of HAND1, RGS16, ACVR2B and RBFOX2 were assessed using qPCR. B2M was used as a housekeeping gene. The graph plots gene expression (dCt expression) relative to B2M and is representative of three independent experiments. The significance was determined using one-way ANOVA with Dunnett’s multiple comparisons test (n = 3, *P-value ≤ 0.05). (B) HEK293T and MDA-MB-231 cells were co-transfected with NT or let-7b-5p mimic or inhibitor, and HAND1 or RGS16 reporter plasmid encoding either WT or mutated 3′ UTR. Twenty-four hours later, the cell lysates were prepared and subjected to dual luciferase assay to measure both firefly (control) and renilla luciferase (experimental signal). The graph plots fold-change relative to NT control and is representative of three independent experiments. The significance was determined using two-way ANOVA with Sidak’s multiple comparisons test (n = 3, *P-value ≤ 0.05). (C) KELLY cells were transfected with siRNAs against MYCN, LIN28B, HAND1, RGS16, ACVR2B, RBFOX2 or NT control in 384-well format. The images of the wells were taken every 2 h for 3 days using IncuCyte ZOOM system. Cell viability was determined using CellTiter-Glo luminescent assay. Both cell confluence and cell viability were plotted as values normalized to the NT control, and each graph is a representative of three independent experiments. The significance was determined using one-way ANOVA with Dunnett’s multiple comparisons test (*P-value ≤ 0.05).