Abstract
Real-time quantitative reverse transcription PCR is a sensitive and widely used technique to quantify gene expression. To achieve a reliable result, appropriate reference genes are highly required for normalization of transcripts in different samples. In this study, 9 previously published reference genes (60S, Fbox, ELF1A, ELF1B, ACT11, TUA5, UBC4, G6PD, CYP2) of soybean [Glycine max (L.) Merr.] were selected. The expression stability of the 9 genes was evaluated under conditions of biotic stress caused by infection with soybean mosaic virus, nitrogen stress, across different cultivars and developmental stages. ΔCt and geNorm algorithms were used to evaluate and rank the expression stability of the 9 reference genes. Results obtained from two algorithms showed high consistency. Moreover, results of pairwise variation showed that two reference genes were sufficient to normalize the expression levels of target genes under each experimental setting. For virus infection, ELF1A and ELF1B were the most stable reference genes for accurate normalization. For different developmental stages, Fbox and G6PD had the highest expression stability between two soybean cultivars (Tanlong No. 1 and Tanlong No. 2). ELF1B and ACT11 were identified as the most stably expressed reference genes both under nitrogen stress and among different cultivars. The results showed that none of the candidate reference genes were uniformly expressed at different conditions, and selecting appropriate reference genes was pivotal for gene expression studies with particular condition and tissue. The most stable combination of genes identified in this study will help to achieve more accurate and reliable results in a wide variety of samples in soybean.
Introduction
Real-time quantitative reverse transcription PCR (RT-qPCR) is one of the most commonly used techniques to examine transcript levels due to its sensitivity, specificity, wide dynamic range, and high throughput capacity [1–3]. As a valuable tool for basic research, RT-qPCR is available in many fields, such as diagnostics, biotechnology and microbiology [4–9]. However, the accuracy is affected by many factors, such as the quantity and integrity of RNA samples, the efficiency of reverse transcription, PCR amplification and variations in the initial quantities of RNA [3, 10–12]. To avoid the influence of these factors, it is necessary to select ideal reference gene(s) to normalize RT-qPCR analysis. The reliability of the RT-qPCR result depends on carefully chosen experimental operation, and especially on the choice of reference genes to ensure proper normalization [13].
Common reference genes used in RT-qPCR for normalization are housekeeping genes related to basic metabolism pathway, such as ACTIN (essential for cytoskeleton structuring and kinetics), elongation factors (ELF1α and ELF1β), 60s (60s Ribosomal protein L30), tubulin (α- and β-tubulin, TUA and TUB, essential for cytoskeleton structuring and kinetic), Fbox (Fbox protein family), Ubiquitin-conjugating enzyme E2 (family UBC4-enzyme involved in abnormal and short-lived proteins degradation), glucose-6-phosphate dehydrogenase (G6PD-important enzyme of the glycolysis pathway) and cyclophylin (CYP-central to protein unfolding and protein interaction) [2, 3, 12–15]. As a consensus, it is assumed that these housekeeping genes have constant expression levels among control and treated samples regardless of experimental conditions, developmental stages, tissues and organs or stress treatments [12, 13, 16, 17]. However, a number of studies reported that transcription of housekeeping genes can fluctuate considerably under certain stress conditions, like pathogen infection, cold temperature and drought [18–20]. For example, when maize seeds were infected by fungi, the expression level of some genes involved in metabolism, protein synthesis were down-regulated, including housekeeping genes like GAPDH [18, 20]. Thus, there are no universal reference genes under all experimental conditions [17, 21, 22].
During the past few years, there have been lots of reports published with the aim of identifying and evaluating suitable reference genes for expression analysis in different plant species under abiotic stress [11, 13, 16, 19, 23, 24], at different developmental stages or tissues [1, 3, 12, 21, 22], after infection with fungi, virus or bacteria [3, 13, 17, 18, 20, 24, 25]. However, there is a lack of validated reference genes under soybean mosaic virus (SMV) infection and nitrogen stress. Investigating mechanisms of virus resistance and nitrogen stress of soybean have always been our research focus. To understand the expression patterns of genes response to these stresses, RT-qPCR is becoming conventional and has helped decipher the functions of target genes. In this report, we analyzed the expression stability of nine housekeeping genes under different experimental conditions (SMV inoculation, different developmental stages and nitrogen stress) in soybean. To obtain reliable RT-qPCR results, ΔCt approach and geNorm program were both used to evaluate the stability of the nine candidate reference genes. The primary objective of this study was to determine which reference genes demonstrate high stability under specific conditions.
Materials and methods
SMV inoculation
The SMV strain SC3 was provided by the National Center for Soybean Improvement (NCSI, Nanjing Agricultural University, Nanjing, China) and maintained in leaves of susceptible cultivar Nannong 1138–2 [26]. The resistant cultivar Zhongdou No.32 (ZD32) and susceptible cultivar Zhongdou No.29 (ZD29) were developed by our institute [27, 28]. These three cultivars were planted in a net house. The SMV inoculums were prepared by grinding leaves of SC3-infected cultivar Nannong 1138–2 to slurry with a pestle in a mortar with moderate 0.01 M sodium phosphate buffer (a mixture of sodium phosphate and potassium phosphate, 5 mL/g leaf tissue, pH 7.2). The inoculation experiments were conducted at V1 stage (Vegetative 1-fully developed leaves at unifoliolate node) of development. The newly expanded unifoliate leaves of the three cultivars were inoculated by rubbing with a paintbrush gently. On the same time, leaves inoculated with 0.01 M sodium phosphate buffer were used as non-infected controls. The inoculated leaves were rinsed with tap water after inoculation. Inoculated leaves of the three cultivars and controls were sampled at 15 min and 6h post SMV inoculation, and frozen in liquid nitrogen and kept at -80°C until RNA isolation.
Developmental stages
Two soybean varieties Tanlong No. 1 (TL1) and Tanlong No. 2 (TL2) were chosen based their phenotypes. TL1 is characterized by ovate leaflet shape and low seed number per pod and TL2 characterized by narrow leaflet shape and high seed number per pod. They were planted on the farm of our institute with three replications. Each plot contained 10 rows of 3.3 m long, with 0.4 m between rows and 0.1 m between individual plants. Apical buds of the two cultivars were sampled at three soybean developmental stages: Vegetative E (VE-characterized by the presence of the cotyledons), Vegetative 1 and Vegetative 3 (V3-the third node with fully developed leaves).
Nitrogen stress
Soybean low-N-tolerant soybean variety “Pohuang” (PH) [29] was chosen as the plant material. The mature seeds were germinated and grown hydroponically in one-half-strength modified Hoagland solution containing 2 mM Ca(NO3)2·4H2O, 2.5 mM KNO3, 0.5 mM NH4NO3, 0.5 mM KH2PO4, 1 mM MgSO4·7H2O, 0.05 mM Fe-EDTA, 0.005 mM KI, 0.1 mM H3BO3, 0.1 mM MnSO4·H2O, 0.03 mM ZnSO4·7H2O, 0.0001 mM CuSO4·5H2O, 0.001 mM Na2MO4·2H2O, 0.0001 mM CoCl2·6H2O [29]. Fourteen-day-old soybean seedlings with cut-off cotyledons were transferred to low nitrogen half Hoagland solutions (10% of normal nitrogen concentration) and high nitrogen (10 times of normal nitrogen concentration) respectively [29, 30]. The culture solution was changed every 3 d. The roots and shoots were sampled separately after 4 h and 6 d of the nitrogen treatment, with three biological replicates per sample. Soybean seedlings in half Hoagland solution without nitrogen treatment were used as controls. The plant tissues were frozen in liquid nitrogen immediately and kept at -80°C until RNA isolation.
RNA isolation and cDNA synthesis
In all treatments above, samples were collected from 5–10 plants and pooled together. Total RNA was extracted from these samples using TRIZOL reagent (Invitrogen, USA) following the procedures provided by the manufacturer. The quantity of RNA was evaluated by electrophoresis on a 1.2% agarose gel, and the concentration was measured by an Epoch microplate spectrophotometer (BioTek, USA). To eliminate any possible DNA contamination, RNA samples were treated with RNase-free DNase I (Thermo Scientific, USA) according to the manufacturer's instructions. First-stand cDNA was synthesized from 1 μg RNA using the PrimeScript reagent Kit with gDNA Eraser (Takara) in a 20 μL reaction according to the supplier’s protocol. The cDNA samples were stored at -20°C for further analysis.
RT-qPCR and data analysis
We selected nine candidate reference genes based on previous studies: 60S, Fbox, ELF1A, ELF1B, ACT11, TUA5, UBC4, G6PD and CYP2 [14, 19, 30, 31], as shown in Table 1. The primer specificity was confirmed by electrophoresis the products of amplification through a 1.2% agarose gel, and all PCR products revealed the presence of the expected amplicons. The quantitative real-time PCR for gene expression was performed on the Bio-Rad thermal using 1×SYBR Green SuperReal Premix (Tiangen, China). Each 20 μL reaction volume contained 4 μL cDNA, 10 μL 1×SYBR Green SuperReal Premix, 4.8 μL dH2O and 0.6 μL each primer. The reaction conditions included an initial denaturation step of 95°C for 15 min, followed by 40 cycles at 95°C for 10 s, 60°C for 32 s and 72°C for 30 s with fluorescent signal recording. All experiments were performed in experimental triplicates and biological duplicates. Threshold cycle (CT) data were collected automatically by software supplied with Bio-Rad thermal cycler. Background-corrected raw fluorescence data were exported from the Bio-Rad thermal cycler and analyzed in LinRegPCR software to calculate the amplification efficiency [32]. Mean CT and standard deviations of the 9 candidate reference genes under different stress treatments were calculated (S2 Table) for further analysis.
Table 1. List of primer sequence and related information for each candidate reference gene.
| Gene | Function | Locus name | Primer sequence | Amplicon length (nt) | Amplification Efficienciesa |
References |
|---|---|---|---|---|---|---|
| 60S | 60s Ribosomal protein L30 | Glyma17g05270 | AAAGTGGACCAAGGCATATCGTCG | 125 | 1.868 | [19] |
| TCAGGACATTCTCCGCAAGATTCC | ||||||
| Fbox | F-box protein family | Glyma12g05510 | AGATAGGGAAATTGTGCAGGT | 93 | 1.865 | [19] |
| CTAATGGCAATTGCAGCTCTC | ||||||
| ELF1A | Eukaryotic elongation factor 1 α | Glyma19g07240 | GACCTTCTTCGTTTCTCGCA | 195 | 1.961 | [14,19,31] |
| CGAACCTCTCAATCACACGC | ||||||
| ELF1B | Eukaryotic elongation factor 1 β | Glyma02g44460 | GTTGAAAAGCCAGGGGACA | 118 | 1.933 | [14,19,31] |
| TCTTACCCCTTGAGCGTGG | ||||||
| ACT11 | Actin | Glyma18g52780 | ATCTTGACTGAGCGTGGTTATTCC | 126 | 1.924 | [30] |
| GCTGGTCCTGGCTGTCTCC | ||||||
| TUA5 | α-tubulin | Glyma05g29000.1 | AGGTCGGAAACT CCTGCTGG | 159 | 1.915 | [14,31] |
| AAGGTGTTGAAGGCGTCGTG | ||||||
| UBC4 | Ubiquitin-conjugating enzyme E2 | Glyma18g44850 | GAGCGAGCAGTTTCAGAC | 168 | 1.910 | [31] |
| CATAGGAGGGACGATACG | ||||||
| G6PD | Glucose-6-phosphatedehydrogenase | Glyma19g24250 | ACTCCTTGATAC CGTTGTCCAT | 126 | 1.931 | [14,31] |
| GTTTGTTATCCGCCTACAGCCT | ||||||
| CYP2 | Cyclophilin 2 | Glyma12g02790 | CGGGACCAGTGTGCTTCTTCA | 154 | 1.853 | [14,19,31] |
| CCCCTCCACTACAAAGGCTCG |
aAs calculated by LinRegPCR software.
Results
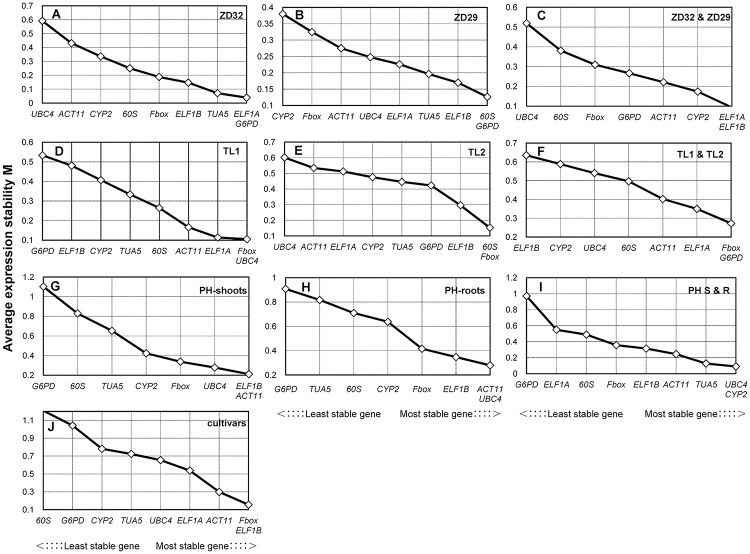
To obtain high accuracy of stability ranking of the 9 candidate reference genes, two different statistical algorithms ΔCt [33] and geNorm (version 3.5) [34] were used to evaluate expression stability of reference genes. The ΔCt algorithm used the mean of standard deviations of delta Cts to rank the performance of each candidate reference gene [33]. The average standard deviations (STDEV) of the 9 genes were calculated respectively (Table 2 and S3 Table). The lower the values are, the more stable the expression of the reference gene is. For geNorm, the approach determined an average expression stability (M) value for each gene to rank the stable level of the candidate reference genes based on their expression stability [3, 34]. It has been shown that M value and gene stability have a negative correlation [12]. Genes with the highest M value are considered to be the least stable ones, while those with the lowest M value have the most stable expression. The geNorm results for all the experimental sets are presented in Fig 1 and S4 Table.
Table 2. Average standard devation (SD) of delta Ct of RT-qPCR.
| SMV inoculation | Developmental stages | Nitrogen stress | cultivars | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZD32 | ZD 29 | 32a & 29b | TL1 | TL2 | 1c & 2d | Shoots | Roots | S & R | |||
| 60S | 0.6441 | 0.3564 | 0.6582 | 0.4778 | 0.4726 | 0.7036 | 1.2416 | 0.8713 | 0.9989 | 1.7921 | |
| Fbox | 0.4076 | 0.5149 | 0.4706 | 0.5299 | 0.5275 | 0.4742 | 0.8117 | 0.8170 | 0.8098 | 0.9796 | |
| ELF1A | 0.4002 | 0.3341 | 0.3928 | 0.4664 | 0.5354 | 0.5866 | — | — | 1.0280 | 0.9867 | |
| ELF1B | 0.4335 | 0.2887 | 0.3978 | 0.6535 | 0.5409 | 0.7765 | 0.7554 | 0.7261 | 0.7438 | 0.9214 | |
| ACT11 | 0.5526 | 0.3433 | — | 0.4777 | 0.5779 | 0.6029 | 0.8051 | 0.8050 | 0.6884 | 0.8905 | |
| TUA5 | 0.4519 | 0.4193 | 0.4104 | 0.5014 | 0.6632 | — | 1.4861 | 1.1665 | 0.7037 | 1.1903 | |
| UBC4 | 1.1844 | 0.3097 | 0.9349 | 0.4616 | 0.8345 | 0.7755 | 0.8110 | 0.8638 | — | 1.0901 | |
| G6PD | 0.4090 | 0.3916 | 0.5097 | 0.7180 | 0.5923 | 0.5127 | 1.9231 | 1.1811 | 2.4452 | 1.7061 | |
| CYP2 | 0.5418 | 0.5733 | 0.3813 | 0.5123 | 0.6667 | 0.6497 | 0.9877 | 0.8317 | 0.6548 | 1.2464 | |
Data obtained for the top five genes are shown in bold letters, while the top three genes are in italic and bold letters.
32a, ZD32;
29b, ZD29;
1C, TL1;
2d, TL2.
Fig 1. Expression stability and ranking of 9 reference genes under SMV treatment as determined by geNorm.
(A) Leaves of ZD32 at 15 min and 6 h post-inoculation with SMV and control. (B) Leaves of ZD29 at 15 min and 6 h post-inoculation with SMV and control. (C) Leaves of ZD32 and ZD29 at 15 min post-inoculation with SMV. (D) Apical buds of TL1 at VE, V1 and V3 stages. (E) Apical buds of TL2 at VE, V1 and V3 stages. (F) Apical buds of TL1 and TL2 at VE and V1 stages. (G) Shoots of PH under nitrogen stress. (H) Roots of PH under nitrogen stress. (I) Shoots and roots of PH under nitrogen stress. (J) Leaves of ZD32 inoculated with SMV, apical buds of TL1 at VE stage and leaves of PH under nitrogen stress.
Expression stability of reference genes under SMV stress
Since the stability of a reference gene is not constant, the most stable genes in one condition can be highly variable in another. Therefore, we analyzed the data based on individual stresses to search for the best reference gene(s) for each stress treatment. Under SMV treatment, results obtained from ΔCt analysis showed that the top five most stably expressed genes were ELF1A, Fbox, G6PD, ELF1B and TUA5 in resistant cultivar ZD32 (Table 2). When the same data were analyzed by geNorm program, the top five genes were ELF1A, G6PD, TUA5, ELF1B and Fbox (Fig 1A). Interestingly, the two methods revealed that the top five most stably expressed genes were the same although the exact order was different, and ELF1A was the best one identified by two methods. For the susceptible cultivar ZD29, when we compared the data obtained by two algorithms, it turn out that the top five most stably expressed genes were overlapped though the order was not the same (Table 2 and Fig 1B). According to geNorm, 60S and G6PD were the most stable reference genes with a combined M value for both genes of 0.126, while CYP2 was the least stable reference gene (M = 0.380) in ZD29 infected with SMV (S4 Table). Values of M that surpassed the cutoff value of 0.5 were not considered stable across the treatments [3]. In general, the M values for the majority of the candidate reference genes were below the cutoff of 0.5, with M scores for a few genes above this value. When considering the resistant and susceptible cultivars, ELF1A and ELF1B behaved best and were the most stable reference genes (M = 0.093) identified by geNorm program (Fig 1c and S4 Table), whereas when using ΔCt method, CYP2 was the most stable gene with the lowest STDEV value, followed by ELF1A and ELF1B.
Expression stability of reference genes at different developmental stages
To confirm the reliability of the potential reference genes at different developmental stages, the validation of the reference genes in TL1 and TL2 was performed. Both the ΔCt method and geNorm algorithm showed similar results, which strengthened the legitimacy of the obtained results. In the developmental series, UBC4, ELF1A and ACT11 were the most stable reference genes in TL1 by ΔCt analysis. Similarly, according to geNorm algorithm, UBC4 and ELF1A were still the top ranked reference genes, with M scores of 0.105 and 0.114, respectively [S4 Table]. In TL2 the ΔCt method and geNorm algorithm both ranked 60S/Fbox as the most stable reference gene pair at different developmental stages. When we compared the two cultivars, the top four most stably expressed genes identified by ΔCt and geNorm were Fbox, G6PD, ELF1A and ACT11, and even the order were exactly the same (Table 2 and Fig 1D–1F).
Expression stability of reference genes under nitrogen stress
We next searched for the best reference genes among the 9 selected candidate reference genes for gene expression analysis under nitrogen stress in shoots and roots of PH, respectively. The top five genes in shoots under nitrogen stress were identical via the two algorithms (Table 2 and Fig 1G–1I), of which ELF1B and ACT11 were the most stable genes among all tested genes, while G6PD remained to be the least stable one. In roots, ELF1B and ACT11 are the top two stable genes identified by ΔCt algorithm, and they are also included in the top 3 stable genes detected by geNorm software (Table 2, Fig 1H). Finally, when different tissues were considered for stability analysis, ΔCt method showed that the most stably expressed gene was CYP2, followed by ACT11 and TUA5 under nitrogen stress (Table 2). However, based on the geNorm results, the order of the three best reference genes was as follows: UBC4, CYP2 and TUA5 (Fig 1I).
Expression stability of reference genes among three cultivars
The stability of reference genes was dissected in various samples under corresponding stresses, including inoculated leaves of ZD32, apical buds of TL1 at VE stage and shoots of PH under high N stress for 4 h. geNorm results indicated that the most stable genes were Fbox and ELF1B with a combined M values of 0.155, followed by ACT11 with 0.299, ELF1A with 0.537 and UBC4 with 0.655 (Fig 1J and S4 Table). When the data were analyzed by ΔCT algorithm, the top five genes were the same (Table 2 and Fig 1J) though the ranking order of expression stability was distinct. The Fbox gene was the most stable one when analyzed by geNorm, while it is the third stable reference gene when ΔCt method was used.
Optimal number of reference genes for normalization
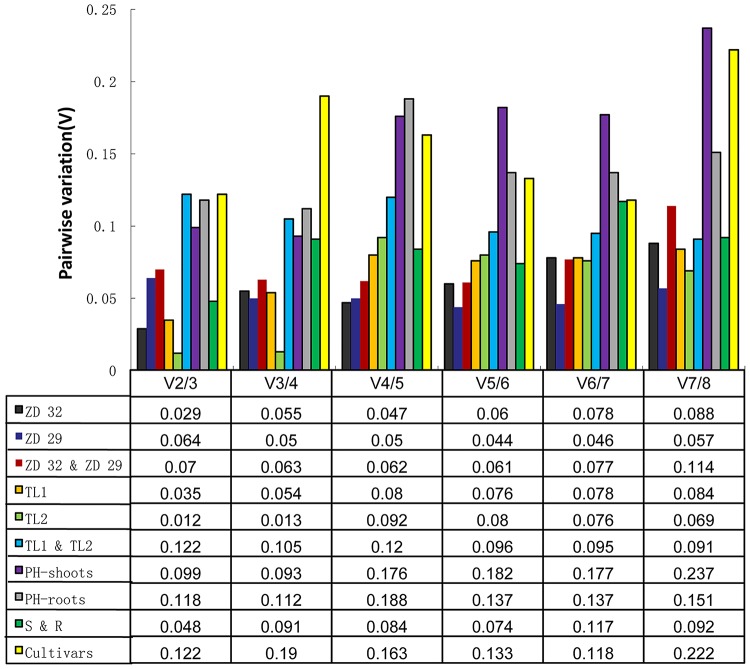
The pairwise variations (Vn/Vn+1) were also calculated with geNorm between two sequential ranked genes to determine the optimal number of reference genes for normalization under a given set of experimental condition. As suggested by Vandesompele et al. [34], a threshold of 0.15 was set as the cut-off value, below which an additional reference gene was not needed. For example, V2/3<0.15 means that the combination of two most stable reference genes was sufficient to normalize the expression of target genes. When considering SMV treatments (ZD32, V2/3 = 0.029; ZD29, V2/3 = 0.064; ZD32 & ZD29, V2/3 = 0.07), different developmental stages (TL1, V2/3 = 0.035; TL2, V2/3 = 0.012; TL1 & TL2, V2/3 = 0.122), nitrogen stress (Shoots, V2/3 = 0.099; Roots, V2/3 = 0.118; Shoots & Roots, V2/3 = 0.048), and different cultivars (ZD 32 & TL1 & PH, V2/3 = 0.122) (Fig 2), the V2/3 values of all the experimental sets were all lower than the cut-off of 0.15, indicating that it is sufficient to use two reference genes for accurate normalization.
Fig 2. Gene expression pairwise variation (V) of the candidate reference genes calculated by geNorm.
The pairwise variation (Vn/n+1) was analyzed between the normalization factors NFn and NFn+1 by geNorm program to determine the optimal number of reference genes required for effective normalization of RT-qPCR data.
Recommended reference genes for RT-qPCR in soybean
In present study, two mathematical and statistical models, ΔCt algorithm and geNorm program were used to determine the most suitable reference genes. With comprehensive analyses with geNorm and ΔCt results, we proposed the most suitable gene pairs for normalization of the target genes under specific experimental conditions (Table 3). For example, the pairwise variation V2/3 value in TL1 was calculated to be 0.035 by geNorm (S4 Table), suggesting that two most stable genes (Fbox and UBC4) can be selected for normalization. However, with ΔCt approach, UBC4 was the most stable gene, ELF1A was the second (ranked the third by geNorm) and the Fbox gene was the seventh. Therefore, UBC4 and ELF1A would be suitable for RT-qPCR in TL1. Analogous analysis was made for each experimental setting (Table 3). Although we could not identify any single gene expressed constantly under all experimental conditions, one or two appropriate stable reference genes in specific given conditions used in RT-PCR experiments could be recommend. And it should guide the selection of reference genes for gene expression analysis in soybean.
Table 3. Recommended reference genes for RT-qPCR of soybean as predicted by geNorm and ΔCt algorithms.
| Recommend genes | ||
|---|---|---|
| SMV stress | ZD32 | ELF1A/G6PD |
| ZD29 | ELF1A /ELF1B | |
| ZD32 & ZD29 | ELF1A/ELF1B | |
| or CYP2/ ELF1A | ||
| or CYP2/ ELF1B | ||
| Developmental stages | TL1 | UBC4/ELF1A |
| TL2 | 60S/Fbox | |
| TL1 & TL2 | Fbox/G6PD | |
| Nitrogen stress | Shoots | ELF1B/ACT11 |
| Roots | ACT11/ELF1B | |
| S & R | CYP2/ACT11 | |
| or CYP2/TUA5 | ||
| cultivars | ZD 32 & TL1 & PH | ELF1B/ACT11 |
Discussion
In RT-qPCR analysis, it is assumed that reference genes have constant expression levels among different samples. However, evidences showed that transcripts levels of housekeeping genes may vary considerably under experimental conditions and/or in tissues types [2]. To obtain high accuracy, it is necessary to validate reference genes for each plant species being studied and for each specific experimental condition [3]. The target gene expression was evaluated according to reference gene expression level, thus unstable reference genes can result in inaccurate evaluation of target gene expression. As reported by Dung et al. in the previous study, the expression of four known dehydration-inducible genes GmNACs in dehydrate treatment was hard to detect with the least stable reference gene of SUBI2, while the relative transcript abundance of GmNACs was induced by 3- to 4-fold with the stable reference genes of 60S and Fbox [19]. A previous report also described that when the two most stable genes GAPDH1 and EF were used as reference genes for pistillate flower, the expression level of AGAMOUS gene increased gradually in stage 1 to stage 4 and then declined at stage 5. However, when the least stable reference gene PLA was used, the expression level of AGAMOUS showed fluctuations and failed to achieve a consistent expression pattern [1]. Under salinity or drought stress, the expression of salinity and drought response gene FaWRKY in roots of tall fescue peaked at 3 h post salt stress with the most stable reference genes (SAND and TUB) used, however, the expression of FaWRKY1 did not show a consistent pattern with the least stable gene (EF1α) used [35]. These data indicated that use of suitable internal controls could reveal a more reliable result and is critical for RT-qPCR analysis. Thus, the selection of suitable reference genes in RT-qPCR analysis is pivotal to normalize the transcript expression of target genes.
The algorithms geNorm and ΔCt have been successfully employed to determine the stability of reference gene expression and identify stable reference genes for various plants species [1]. The results obtained from both methods were similar in most of the analyses. For example, ELF1B and ACT11 were found to be the most stable reference genes among different soybean cultivars identified by two ways (Fig 1J, Table 3). In shoots, the top five most stably expressed genes, even their order was exactly the same according to the two methods under nitrogen stress (Fig 1G, Table 2). Some inconsistencies were also found in the ranking order between the two statistical analytical programs, which may be caused by distinct statistical algorithm procedures. In our study, when ZD29 infected with virus, G6PD was the best internal control identified by geNorm software, but only ranked the sixth by ΔCt approach and may be inappropriate to normalize in RT-qPCR. Different algorithms strategies may lead to different selection of suitable reference genes, which consistent with previous studies [1, 12, 19, 24, 25]. Thus, the combination of two or more analysis methods to determine the most accurate reference genes for different treatment conditions is necessary.
It has been suggested that the number of reference genes required for quantifying gene expression should depend on the consideration of the research purpose. To get a rough expression of the target gene, one most stable reference gene may be enough, whereas two or more reference genes must be taken if a more accurate expression level was needed [2, 14, 34, 36]. The optimal number of reference genes should be decided based on the threshold of 0.15, nevertheless it is not absolute since small datasets require fewer reference genes than larger ones [12]. In our study, under SMV treatment, the values of V2/3–V7/8 were totally below 0.15 (Fig 2), meaning 2–7 reference genes were feasible. The results also suggested that V value should only be used as reference, but not the judgment criterion.
The nine commonly used reference genes evaluated here were 60S, Fbox, ELF1A, ELF1B, ACT11, TUA5, UBC4, G6PD and CYP2. Similarly to our selected control genes, analogous results were observed in soybean for which ELF1B/60S and 60S/Fbox were thought to be the most stable gene pairs in roots and shoots respectively under various stresses [19]. In previous studies, we also found that, ELF1B exhibited highly stable expression under SMV infection [24] and at different developmental stages [15]. In our study, the gene pairs ELF1A/ELF1B, CYP2/ELF1A or CYP2/ELF1B were the most stable reference genes under SMV treatment among resistant and susceptible cultivars. Similar results were obtained by Vívian et al., who reported that GmCYP2 and GmELF1A genes showed relatively stable expression levels in leaves attacked by soybean caterpillar [31]. The expression stability of CYP was also described by Bansal et al. [25], who found CYP to be a potential stable reference gene during powdery mildew in soybean. It is verified by previous work which showed that Fbox was recommended for use in soybean under the conditions of powdery mildew or aphid, and in Brassica napus under cold stress or salicylic acid treatment [25, 36]. Similarly, in this study, Fbox exhibited stable expression at different developmental stages of TL1 and TL2 (Fig 1D–1F, Table 3).
Although ACT is one of the most commonly used reference gene in plants, its expression may vary considerably between tissues and/or samples even within the same plants. We noted that ACT has been considered a consistent reference gene and ranked as highly effective for gene expression studies with soybean at different lighting periods [24] and various developmental stages [2]. However, in Cycas elongate ACT ranked last indicating low stability across different tissue samples [12]. And the low expression stability was also observed in Jatropha curcas [1], Setaria viridis [16] and Nicotiana tabacum [37]. In present study, ACT11 was the most stable reference gene in both shoots and roots under nitrogen stress (Fig 1G–1I, Table 3), and it also exhibited stable expression among different cultivars (Fig 1J). While under SMV treatment, the expression of ACT11 was stable (Fig 1A and 1B) neither in resistant nor in susceptible cultivar. Taken together, these results indicated that suitable reference genes were highly specific for different plant species and particular experimental setups.
We evaluated the independent effect of different experimental sets on the ranking of the reference genes. In the SMV treatment, results indicated that the effect of virus infusion on the expression levels of ELF1A, ELF1B and CYP2 was less than that on the other reference genes. These three genes may not be involved in any of the signaling processes of plants in virus defense, and could be considered as reference genes for gene expression analysis in response to virus. For nitrogen stress, G6PD was the least stable gene both in root and shoot and should be avoided to be internal control, which was in accordance with the results of other researchers [14, 31, 38], indicating that G6PD was not only a component of the glycolytic pathway but also participated in other biological processes.
We noted that ELF1A has been considered as the most unstable gene in Cycas elongate [12], tomato [18] and tall fescue [35]. While we report the results which are contrary to the previous observations, ELF1A appears to be a reliable reference gene under SMV treatment. Our results corroborate those obtained by previous studies, in which ELF1A has been considered a stable and effective reference gene in gene expression studies with potato [39], Populus [40], poplar [41] and Caragana [42, 43]. In addition, we also found that the expression stability of ELF1A has shown distinct performance in soybean with different experimental conditions [19, 34, 38]. The contrasting results among different species or within the same species presented a differential expression profile of ELF1A, which could be attributed to different treatments. On the other hand, the different results obtained from different studies may be due to the primer pairs that amplify the ELF1A member, in accordance with the previous study on the expression stability of 6 soybean EF1α genes (named EF1α1a1, EF1α1a2, EF1α1b, EF1α2a, EF1α2b and EF1α3), which was proposed by Saraiva et al. [44]. The third, ELF1A may have species diversity in the process of evolution, and its function may have changed among different species. These results provided guidelines for selecting appropriate reference genes in gene expression studies with a particular experimental setting.
Conclusion
In present study, the expression of 9 candidate reference genes under different experimental conditions was evaluated using ΔCt and geNorm algorithms and the most suitable internal controls for normalizing the data of RT-qPCR under specific conditions in soybean were confirmed.
For SMV infection, ELF1A/G6PD and ELF1A/ELF1B should be reliable reference genes for resistant and susceptible cultivars, respectively, and the best reference gene pair is ELF1A/ELF1B between resistant and susceptible cultivars. For different developmental stages, UBC4/ELF1A is the best combination for TL1 and 60S/Fbox for TL2. In addition, Fbox/G6PD should be used as the most suitable reference for comparisons between the two cultivars. For nitrogen stress, ELF1B/ACT11 should be used for the shoots and ACT11/ELF1B for the roots and CYP2/ACT11 is the best reference gene pair between shoot and root tissues. The combination of ELF1B and ACT11 could be used as the best gene pair for comparison among different cultivars under corresponding stress treatment.
Supporting information
(PDF)
(PDF)
(XLS)
(PDF)
Acknowledgments
We thank the National Center for Soybean Improvement of Nanjing Agricultural University for providing SC3 virus strains and advice. We greatly thank the anonymous reviewers for critical reading and suggestions on how to improve the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was supported by the National Natural Science Foundation (Grant No.31371653 and 31401410), http://www.nsfc.gov.cn/publish/portal1/, and the National Transgenic Project of China (Grant No.2014ZX0800930B-003), http://www.nmp.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Karuppaiya P, Yan XX, Liao W, Wu J, Chen F, Tang L. Identification and validation of superior reference gene for gene expression normalization via RT-qPCR in staminate and pistillate flowers of Jatropha curcas—a biodiesel plant. Plos ONE. 2017; 12(2): e0172460 doi: 10.1371/journal.pone.0172460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu R, Fan C, Li H, Zhang Q, Fu YF. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol. 2009; 10(1): 93 doi: 10.1186/1471-2199-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aks F, Tdf S, Romanel E, Msf V. microRNAs as reference genes for quantitative PCR in cotton. Plos ONE. 2017; 12(4): e0174722 doi: 10.1371/journal.pone.0174722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracht AJ, O’Hearn ES, Fabian AW, Barrette RW, Sayed A. Real-time reverse transcription PCR assay for detection of Senecavirus A in swine vesicular diagnostic specimens. Plos ONE. 2016; 11(1): e0146211 doi: 10.1371/journal.pone.0146211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wulfken LM, Moritz R, Ohlmann C, Holdenrieder S, Jung V, Becker F, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. Plos ONE. 2011; 6(9): e25787 doi: 10.1371/journal.pone.0025787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeringer EM, Rai AJ, DeCastro J, Qu L, Gonzalez M, Chapman L, et al. A complete workflow for high throughput isolation of serum microRNAs and downstream analysis by qRT-PCR: application to cancer biomarker discovery. Clin Res. 2015; 75(15 Supplement): 3387. [Google Scholar]
- 7.Chaouachi M, Nabi N, Hafsa AB, Zellama MS, Skhiri F, Saïd K. Monitoring of genetically modified food and feed in the Tunisian market using qualitative and quantitative real-time PCR. Food Sci Biotechnol. 2013; 22(4): 1161–1170. doi: 10.1007/s10068-013-0198-2 [Google Scholar]
- 8.Llanos A, François JM, Parrou JL. Tracking the best reference genes for RT-qPCR data normalization in filamentous fungi. BMC Genomics. 2015; 16(1): 71 doi: 10.1186/s12864-015-1224-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikel P, Vasickova P, Kralik P. Methods for Preparation of MS2 Phage-Like Particles and Their Utilization as Process Control Viruses in RT-PCR and qRT-PCR Detection of RNA Viruses From Food Matrices and Clinical Specimens. Food Environ Virol. 2015; 7(2): 96–111. doi: 10.1007/s12560-015-9188-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Die JV, Román B. RNA quality assessment: a view from plant qPCR studies. J Exp Bot. 2012; 63(17): 6069–6077. doi: 10.1093/jxb/ers276 [DOI] [PubMed] [Google Scholar]
- 11.Li X, Zhang D, Li H, Gao B, Yang H, Zhang Y, et al. Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front Plant Sci. 2015; 6(38): 38 doi: 10.3389/fpls.2015.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Deng T, Chen L, Wu H, Zhang S. Selection and validation of reference genes for qRT-PCR in Cycas elongata. Plos ONE. 2016; 11(4): e0154384 doi: 10.1371/journal.pone.0154384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borges A, Tsai SM, Caldas DGG. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012; 31(5): 827–838. doi: 10.1007/s00299-011-1204-x [DOI] [PubMed] [Google Scholar]
- 14.Jian B, Liu B, Bi Y, Hou W, Wu C, Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol. 2008; 9(1): 1–14. doi: 10.1186/1471-2199-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Libault M, Thibivilliers S, Bilgin DD, Radwan O, Benitez M, Clough SJ, et al. Identification of four soybean reference genes for gene expression normalization. The Plant Genome. 2008; 1: 44–54. doi: 10.3835/plantgenome2008.02.0091 [Google Scholar]
- 16.Martins PK, Mafra V, Souza WRD, Ribeiro AP, Vinecky F, Basso MF, et al. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci Rep. 2016; 6: 28348 doi: 10.1038/srep28348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wieczorek P, Wrzesińska B, Obrępalska-Stęplowska A. Assessment of reference gene stability influenced by extremely divergent disease symptoms in Solanum lycopersicum L. J Virol Methods. 2013; 194(1): 161–168. doi: 10.1016/j.jviromet.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 18.Müller OA, Grau J, Thieme S, Prochaska H, Adlung N, Sorgatz A, et al. Genome-wide identification and validation of reference genes in infected tomato leaves for quantitative RT-PCR analyses. Plos ONE. 2015; 10(8): e0136499 doi: 10.1371/journal.pone.0136499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le DT, Aldrich DL, Valliyodan B, Watanabe Y, Ha CV, Nishiyama R, et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE. 2012; 7(9): e46487 doi: 10.1371/journal.pone.0046487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanubile A, Pasini L, Marocco A. Differential gene expression in kernels and silks of maize lines with contrasting levels of ear rot resistance after Fusarium verticillioides infection. J Plant Physiol. 2010; 167(16): 1398–1406. http://dx.doi.org/10.1016/j.jplph.2010.05.015 [DOI] [PubMed] [Google Scholar]
- 21.Marum L, Miguel A, Ricardo CP, Miguel C. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE. 2012; 7(4): e35113 doi: 10.1371/journal.pone.0035113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C, Ma J, Guo Q, Li X, Wang H, Lu M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE. 2013; 8: e56573 doi: 10.1371/journal.pone.0056573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D, Hu B, Wang Q, Liu H, Pan F, Wu W. Identification and evaluation of reference genes for accurate transcription normalization in safflower under different experimental conditions. Plos ONE. 2015; 10(10): e0140218 doi: 10.1371/journal.pone.0140218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S, Niu H, Liu C, Zhang J, Hou C, Wang D. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS ONE. 2013; 8(10): e75271 doi: 10.1371/journal.pone.0075271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bansal R, Mittapelly P, Cassone BJ, Mamidala P, Redinbaugh MG, Michel A, et al. Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS ONE. 2015; 10(8): e0134890 doi: 10.1371/journal.pone.0134890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Zheng G, Han L, Dagang W, Yang X, Yuan Y, et al. Genetic analysis and mapping of genes for resistance to multiple strains of soybean mosaic virus in a single resistant soybean accession pi 96983. TAG. 2013; 126(7): 1783–1791. doi: 10.1007/s00122-013-2092-y [DOI] [PubMed] [Google Scholar]
- 27.Guo DD, Chen HF, Yang ZL, Shan ZH, Zhu XL, Chen SL, et al. Analysis of resistance inheritance to soybean mosaic virus (SMV) strain SC-3 and mapping related QTL in soybean. Soybean Science. 2012; 4(31): 511–516 [Google Scholar]
- 28.Chen H, Shan Z, Sha A, Wu B, Yang Z, Chen S, et al. Quantitative trait loci analysis of stem strength and related traits in soybean. Euphytica. 2011; 179(3): 485–497. doi: 10.1007/s10681-011-0382-5 [Google Scholar]
- 29.Hao QN, Zhou XA, Sha AH, Wang C, Zhou R, Chen SL. Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genomics. 2011; 12(1): 525 doi: 10.1186/1471-2164-12-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, Hou Y, Hao Q, Chen H, Chen L, Yuan S, et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. Plos ONE. 2015; 10(4): e0125174 doi: 10.1371/journal.pone.0125174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miranda VJ, Coelho RR, Viana AAB, Neto OBO, Carneiro RMDG, Rocha TL, et al. Validation of reference genes aiming accurate normalization of qPCR data in soybean upon nematode parasitism and insect attack. BMC Res Notes. 2013; 6(1): 196 doi: 10.1186/1756-0500-6-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003; 339: 62–66. doi: 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- 33.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006; 7(1): 33 doi: 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, Paepe AD et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3(7): RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Chen Y, Hu B, Tan Z, Huang B. Identification and validation of reference genes for quantification of target gene expression with quantitative real-time PCR for tall fescue under four abiotic stresses. PLoS ONE. 2015; 10(3): e0119569 doi: 10.1371/journal.pone.0119569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Chen Y, Fang H, Shi H, Chen K, Zhang Z, et al. Selection of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in brassica napus, under various stress conditions. Mol Genet Genomics. 2014; 289(5): 1023–1035. doi: 10.1007/s00438-014-0853-1 [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Shi L, Han C, Yu J, Li D, Zhang Y. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One. 2012; 7(9): e46451 doi: 10.1371/journal.pone.0046451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Yu K, Poysa V, Shi C, Zhou Y. Selection of reference genes for normalization of qRT-PCR analysis of differentially expressed genes in soybean exposed to cadmium. Mol Biol Rep. 2012; 39(2): 1585–1594. doi: 10.1007/s11033-011-0897-9 [DOI] [PubMed] [Google Scholar]
- 39.Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005; 56: 2907–2914. doi: 10.1093/jxb/eri285 [DOI] [PubMed] [Google Scholar]
- 40.Xu M, Zhang B, Su X, Zhang S, Huang M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem 2011; 408(2): 337–339. doi: 10.1016/j.ab.2010.08.044 [DOI] [PubMed] [Google Scholar]
- 41.Basa B, Solti Á, Sárvári É, Tamás L. Housekeeping gene selection in poplar plants under Cd-stress: comparative study for real-time PCR normalisation. Funct Plant Biol. 2009; 26: 1079–1087. doi: 10.1071/FP09073 [DOI] [PubMed] [Google Scholar]
- 42.Yang Q, Yin J, Li G, Qi L, Yang F, Wang R. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol Biol Rep. 2014; 41: 2325–2334. doi: 10.1007/s11033-014-3086-9 [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Zhang L, Li W, Han S, Yang W, Qi L. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PloS ONE. 2013; 8(1): e53196 doi: 10.1371/journal.pone.0053196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraiva KD, Fernandes dM D, Morais VD, Vasconcelos IM, Costa JH. Selection of suitable soybean EF1α genes as internal controls for real-time PCR analyses of tissues during plant development and under stress conditions. Plant Cell Rep. 2014; 33(9): 1453–1465. doi: 10.1007/s00299-014-1628-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(XLS)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.