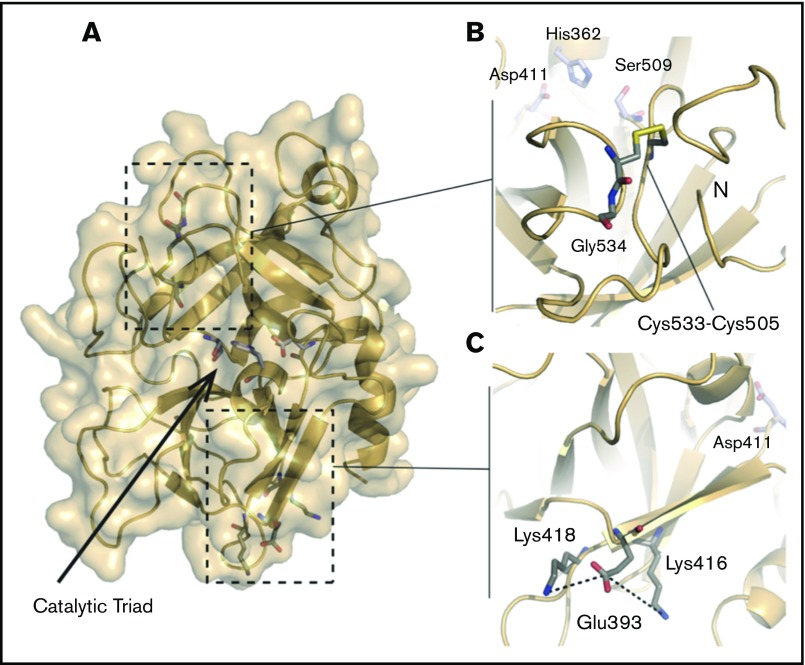
Figure 5.
Structure of the HABP2 protein and locations of C533, E393, and the active site residues. The protein model of HABP2 was constructed using MODELER 9v7, with the crystal structure of the homologous hepatocyte growth factor activator (PDB code: 1YC0) as a template.109 (A) The active site of wild-type HABP2 contains a catalytic triad of 3 residues (E411, H362, and S509, shown in blue and red). (B) C533 forms a cysteine bridge with C505, adjacent to G534 (the site of the Marburg I polymorphism, G534E). These 3 residues are located on the same functionally important surface loop, near the active site residue S509 and the N terminus of the protease domain. The C533F mutation is predicted to break this cysteine bridge, destabilizing these interactions and presumably reducing protein activity. (C) E393 interacts with nearby lysine residues K416 and K418, located on the same β-strand as D411 (part of the catalytic triad). The Marburg II polymorphism (E393Q) is predicted to disrupt this interaction.