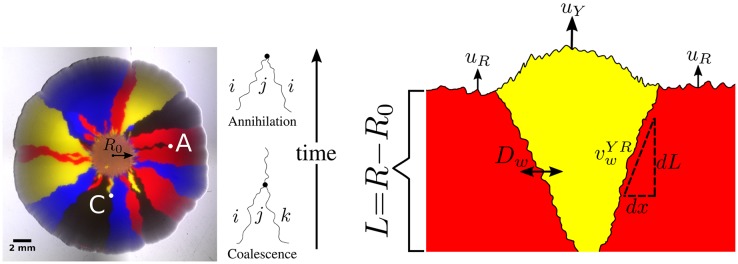
Fig 1.
Left: A four-color E. coli range expansion. Four strains of E. coli differing only by a heritable fluorescent marker were inoculated on an agar plate in a well-mixed droplet and expanded outwards, leaving behind a “frozen record” of their expansion. Virtually all growth occurred at the edge of the colony. The markers instilled different expansion velocities: our eCFP (blue) and eYFP (yellow) strains expanded the fastest, followed by our black strain, and finally our mCherry (red) strain. As a result of the differing expansion velocities, the yellow/blue bulges at the frontier are larger than the black bulges which are larger than the red bulges, although the significant stochastic undulations at the front mask their size. The microbes segregate into one color locally at a critical expansion radius R0 due to extreme genetic drift at the frontier [8]. After segregated domains form, genetic domain walls diffuse and collide with neighboring walls in an “annihilation” or “coalescence” event indicated by an A or C, respectively. Right: Illustration of the relevant parameters used to model range expansions. Here, a faster expanding, more fit yellow strain with expansion velocity uY is sweeping through a less fit red strain with expansion velocity uR, in a regime where the curvature of the colony can be neglected. The length expanded by the colony is L = R − R0. We characterize domain wall motion per differential length expanded dL and the wall’s differential displacement perpendicular to the expansion direction dx. is a dimensionless speed, characterizing the yellow-red (YR) domain wall’s average expansion dx per length expanded dL, i.e., Dw is the domain walls’ diffusion coefficient per length expanded; it controls how randomly the domain walls move. We treat the dynamics of our four strains as a one-dimensional line of annihilating and coalescing random walkers using the parameters R0, Dw, and , where ij represents all possible domain wall types.