Abstract
Background
Sustained inflammation in the heart is sufficient to provoke left ventricular (LV) dysfunction and LV remodeling. Although inflammation has been linked to many of the biologic changes responsible for adverse LV remodeling, the relationship between inflammation and protein quality control in the heart is not well understood.
Methods and Results
To study the relationship between chronic inflammation and protein quality control we utilized a mouse model of dilated cardiomyopathy driven by cardiac restricted overexpression of TNF (Myh6-sTNF). Myh6-sTNF mice develop protein aggregates containing ubiquitin tagged proteins within cardiac myocytes related to proteasome dysfunction and impaired autophagy. The 26S proteasome was dysfunctional despite normal function of the core 20S subunit. We found an accumulation of autophagy substrates in Myh6-sTNF mice, which was also seen in tissue from patients with end stage heart failure. Moreover, there was evidence of impaired autophagosome clearance following chloroquine administration in these mice indicative of impaired autophagic flux. Finally, there was increased mTORC1 activation, which has been linked to inhibition of both the proteasome and autophagy.
Conclusion
Myh6-sTNF mice with sustained inflammatory signaling develop proteasome dysfunction and impaired autophagic flux that is associated with enhanced mTORC1 activation.
Keywords: autophagy, proteasome, inflammation, ubiquitin
Subject Codes: Basic Science Research, Inflammation, Myocardial Biology, Heart Failure, Inflammatory Heart Disease
INTRODUCTION
Sustained inflammatory signaling in the heart leads to the development of a cardiomyopathy characterized by myocardial fibrosis, left ventricular (LV) dilation, and LV dysfunction. If, the inciting cause for the inflammation is resolved (e.g. viral myocarditis), the cardiomyopathy is often fully reversible, even though the extent of LV dysfunction may have been quite severe. Two recent observations suggested the intriguing possibility that impaired protein quality control may play a role in the development of reversible inflammation-induced cardiomyopathy. First, we observed the accumulation of protein aggregates in two different transgenic mouse models of sustained inflammation due to myocyte restricted expression of either tumor necrosis factor (TNF) or TNF receptor associated factor 2 (TRAF2).1, 2 Interestingly mice with conditional expression of TRAF2 develop protein aggregates that are subsequently cleared once inflammatory signaling is switched off.3 Second, bioinformatic analyses of the changes in gene expression in the conditional TRAF2 model suggested that genes involved in the ubiquitin proteasome system and autophagy, the two pathways responsible for protein quality control, were among the most heavily regulated following resolution of inflammatory signaling.3
Although inflammation plays a well-recognized role in regulating the turnover of proteins in skeletal muscle in the setting of atrophy, the role of inflammation with respect to protein quality control in the heart is less well-understood.4 Accordingly, here we sought to evaluate two of the major biological systems involved in the maintenance of protein quality control, namely the ubiquitin-proteasome and the autophagy-lysosome system in a well characterized transgenic model of sustained inflammatory signaling (Myh6-sTNF mice) that develops progressive LV remodeling and LV dysfunction. Here we show for the first time that sustained inflammatory signaling secondary to cardiac restricted overexpression of TNF results in impaired protein quality control secondary to proteasome dysfunction and impaired autophagic flux.
METHODS
Myh6-sTNF mice
The generation and characterization of the transgenic mouse line with cardiac restricted overexpression of secreted TNF (Myh6-sTNF [previously referred to as MHCsTNF]) used in this study is described elsewhere.5 Littermate control mice and Myh6-sTNF mice were studied at 4, 8, or 12 weeks of age, as specified. All animal studies were conducted in accordance with the guidelines of the Animal Studies Committee at Washington University School of Medicine and Baylor College of Medicine Animal Care and Research Advisory Committee.
Ubiquitin-Protein Conjugates
Whole hearts from littermate control and Myh6-sTNF mice were harvested, weighed and then frozen rapidly. Myocardial protein extracts were prepared as described previously,6 and the aliquots of the supernatants were stored at −80 °C. The myocardial protein concentration was determined by the BCA Protein Assay Kit (Pierce, Rockford IL) using serum albumin as a standard. Equivalent amounts of protein extracts from the littermate control and Myh6-sTNF mouse hearts were separated on a 12% SDS polyacrylamide gel under reducing conditions, and then transferred to a nitrocellulose membrane in the presence of 0.02% SDS and 25% methanol at 4 °C (30V over 17hrs). The membrane was autoclaved and then blocked in Tris-buffered saline supplemented with 20% heat inactivated fetal calf serum and 0.45% Tween 20. Western blotting was performed using a rabbit anti-ubiquitin polyclonal antibody (Affiniti-Research Products Limited, Exceter, UK). The primary antibody was visualized by a horseradish peroxidase (HRP) labeled secondary antibody and the ECL chemiluminescence assay (both from Amersham Pharmacia Biotech, Piscataway, NJ). The resulting films (HyperfilmTM ECLTM, Amersham Pharmacia) were scanned on a personal densitometer S1 (Molecular Dynamics, Sunnyvale, CA), and band density (in arbitrary units) were evaluated using ImageQuaNT 4.2a (Molecular Dynamics).
Immunofluoresence microscopy was performed on tissue from 12 weeks old Myh6-sTNF and littermate control mice using laser confocal microscopy (Zeiss Confocal LSM 700). The tissue was fixed in 4% paraformaldehyde for 24 hours at 4 °C and then dehydrated in 70% ethanol. Tissues were embedded in paraffin and cut. Sections were then deparaffinized and rehydrated followed by antigen retrieval using Diva Decloaker solution (Biocare Medical). Slides were blocked in 5% normal donkey serum (Santa Cruz, SC-2044). Primary antibodies were ubiquitin (Abcam, ab134953), p62 (Progen, GP62-C), and desmin (Santa Cruz, sc-7559). Alexa Fluor-conjugated donkey antibodies (Invitrogen) were used as the secondary antibodies.
Proteasome Function
Hearts from Myh6-sTNF and littermate control mice were homogenized in a buffer containing 50 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 250 mM sucrose, 2 mM DTT, and 2 mM ATP, using a 2-ml PYREX tissue grinder. Debris was removed by centrifugation at 10,000 × g for 15 minutes. The 20S and 26S proteasome were isolated after ultra-centrifugations (100,000 × g for 1 hour and 100,000 × g for 5 hours), as described by Solomon and Goldberg.7 Chymotrypsin-like proteasome activity was measured as the release of 7-amino-4-methylcoumarin (AMC) from the fluorogenic peptide substrate suc-LLVY-AMC (Biomol). The change in fluorescence kinetics was measured for 1 hour at 37 °C using a FLx800 Microplate Fluorescence Reader (excitation: 360/40 nm, emission: 530/20 nm; Bio-Tek Instruments, Winooski, VM). The assays were performed in 50 mM Tris-HCl (pH 7.8), 2 mM MgCl2, and 250 mM sucrose, using 10 μg of protein for each reaction. The activity of the 26S proteasome was determined in the above buffer supplemented with 0.25 mM ATP, whereas 20S activity was measured in the presence of 0.01% SDS.
Western blot analyses were performed to determine the protein levels of the subunits of the 19S and 20S proteasome using myocardial protein extracts from 12 weeks old littermate control and Myh6-sTNF mice. Proteasome subunits were quantified using antibodies from Biomol, Plymouth Meeting, PA.
Autophagy
Whole hearts were harvested from 12 weeks old Myh6-sTNF mice and littermate controls. Ventricles were divided into two pieces and rapidly frozen in liquid nitrogen and stored at −80 °C prior to isolation of protein and mRNA. Preparation of protein extracts and Western blotting were performed as previously described.8 The following antibodies were used to assess autophagy substrates: LC3 (Novus Biologicals, NB100-2220), p62 (Abcam, ab56416), GAPDH (Abcam, ab22555), HRP-linked anti-mouse IgG (Jackson ImmunoResearch, 715-035-150), and HRP-linked anti-rabbit IgG (Jackson ImmunoResearch, 715-035-152). Band density was determined using Image Station 4000R Pro (Kodak) and Image J software was used for quantitative analysis.
RNA was isolated using Trizol (Ambion) and the PureLink RNA Mini Kit (Invitrogen). cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative RT-PCR was performed on an ABI 7200 machine (Applied Biosystems). Reactions were performed using Power SYBR Green Master Mix (Applied Biosystems). Predesigned primers for SYBR green based assays were obtained from Integrated DNA Technologies.
To measure autophagic flux Myh6-sTNF mice and littermate controls were treated with chloroquine (Sigma) 40 mg/kg by intraperitoneal (IP) injection to inhibit lysosome function or equal volume of vehicle (normal saline) 4 hours prior to sacrifice. Protein extracts were prepared from whole ventricles and LC3 and GAPDH protein levels were determined by Western blotting as described above.
Lysosome abundance and mTORC1 activation
Lysosome abundance and mTORC1 activation was assessed in tissue harvested from 12 weeks old Myh6-sTNF mice and littermate controls by Western blotting and quantitative RT-PCR as described above. Lysosome abundance was assessed by quantifying mRNA and protein levels for the lysosome markers LAMP1 and LAMP2. The following antibodies were used: LAMP1 (Abcam, ab24170), LAMP2 (Developmental Studies Hybridoma Bank, ABL-93), GAPDH (Abcam, ab22555), HRP-linked anti-rabbit IgG (Jackson ImmunoResearch, 715-035-152), and HRP-linked anti-rat IgG (Cell Signaling, 7077). Predesigned primers for quantitative RT-PCR were obtained from Integrated DNA Technologies. Cathepsin activity was measured as previously described.9 Briefly heart tissue was homogenized in 100 mm sodium acetate pH 5.5, 2.5 mm EDTA, Triton X-100 0.01%, and 2.5 mm DTT. For cathepsin B activity the supernatant was incubated with 100 uMof Z-Arg-Arg-AMC at pH 6.0. For cathepsin D the supernatant was incubated with 10 M substrate7-methoxycoumarin-4-acetyl (Mca)-Gly-Lys-Pro-Ile-Leu-Phe-Phe-ArgLeu-Lys-2,4nitrophenyl (Dnp)-D-Arg-NH2. The fluorescent intensity of the cleavage products was measured using a TECAN Infinite M200 Pro microplate reader.
mTORC1 activation was determined by assessing the ratio of phosphorylated to total protein levels of the downstream targets 4E-BP1 and p70S6K using the following antibodies: phospho-4E-BP1 (Cell Signaling, 9451), total 4E-BP1 (Cell Signaling, 9644), phospho- p70S6K (Cell Signaling, 9234), total p70S6K (Cell Signaling, 9644), and HRP-linked anti-rabbit IgG (Jackson ImmunoResearch, 715-035-152).
Endoplasmic Reticulum Stress
ER stress was measured in tissue harvested from 12 weeks old Myh6-sTNF mice and littermate controls. Western blot was performed as described above using the following antibodies: CHOP (ThermoFisher Scientific, MA1-250), ATF4 (Abcam, ab184909), ATF6 (Cell Signaling, 65880), and Actin (Sigma, A2066). Quantitative RT-PCR for ddit3, sxbp1, and gapdh was performed as described above using predesigned primers from IDT.
Human Myocardial Samples
Human LV tissue from patients with dilated cardiomyopathy that was harvested at the time of cardiac transplant and controls was obtained from the Washington University Translational Cardiovascular Biobank & Repository. Details regarding the clinical data for these human hearts are summarized in the Data Supplement. The procedures for tissue acquisition have been described previously.10 Studies were conducted in accordance with protocols approved by the Washington University School of Medicine IRB and all subjects gave informed consent. Immunofluorescent confocal imaging of p62, ubiquitin, and desmin and Western blotting for p62, LC3, and GAPDH were performed as described above.
Statistical Analysis
All data are expressed as mean ± SEM. Two-way analysis of variance (ANOVA) was used to test for mean differences between littermate control and Myh6-sTNF hearts at 4, 8, and 12 weeks of age. Where appropriate post-hoc analysis of variance testing was performed using a Bonferoni analysis. An unpaired Student’s t test was used to test for mean differences between littermate control and Myh6-sTNF hearts at a single point in time. A difference was said to be significant at the P < 0.05 level.
RESULTS
Accumulation of Ubiquitinated Protein Conjugates in Myh6-sTNF Mouse Hearts
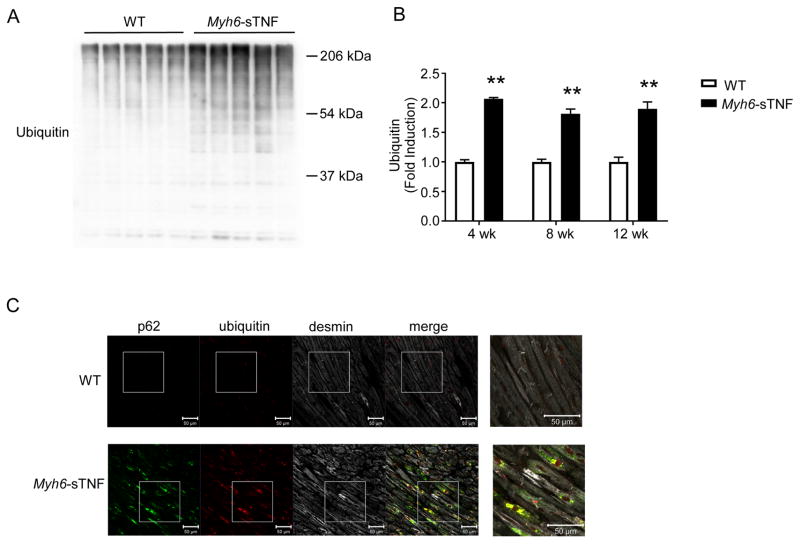
We have previously shown that protein aggregates accumulate in the hearts of mice with cardiac restricted overexpression of TNF, as well as mice with cardiac restricted overexpression of TRAF2 that phenocopy the heart failure phenotype observed in the Myh6-sTNF mice.1, 2 To further assess proteotoxic stress in this setting, we first sought to determine whether there was an accumulation of ubiquitin tagged proteins in the Myh6-sTNF mouse hearts. Figure 1A depicts a representative Western blot of myocardial samples obtained from littermate control and Myh6-sTNF mice at 12 weeks of age. The salient finding shown by Figure 1A is, the amount of ubiquitin-protein conjugates were greater in Myh6-sTNF hearts when compared to littermate controls. The Western blots of myocardial extracts from mice at 4 and 8 weeks of age showed similar findings; that is, the levels of ubiquitin-protein conjugates were elevated significantly in Myh6-sTNF hearts compared to littermate controls (Figure 1B).
Figure 1.
Ubiquitin-protein conjugates in Myh6-sTNF mouse hearts. A) Western blot analysis of ubiquitin-protein conjugates in myocardial extracts from Myh6-sTNF and WT littermate control mice at the age of 12 weeks. B) Group data summarizing the level of ubiquitinated protein conjugates in myocardial extracts from Myh6-sTNF and littermate control mice at the ages of 4, 8, and 12 weeks (n=5 per group). **P <0.001 by 2-way ANOVA with Bonferoni correction. C) Accumulation of protein aggregates containing ubiquitin tagged substrates and p62 in Myh6-sTNF hearts. Representative immunofluorescent images of LV myocardium from WT and Myh6-sTNF mice stained for ubiquitin, p62, and the myocyte specific intermediate filament desmin (N=5 per group). 20× objective, 2.5× zoom on magnified merged image on far right.
To determine the localization of the ubiquitin tagged proteins, we performed immunofluorescence microscopy in 12 weeks old Myh6-sTNF mouse hearts. Confocal imaging demonstrated that the ubiquitinated proteins were incorporated into protein aggregates within individual cardiac myocytes (Figure 1C) in Myh6-sTNF mice, but were not detectable in WT mice. In other disease processes protein aggregates have been shown to contain the adaptor protein p62, which binds to ubiquitin bound proteins and targets them for incorporation into to autophagosomes for subsequent breakdown by the autophagy-lysosome pathway.11 We found that protein aggregates in our model also contained p62. Indeed, desmin, an intermediate filament protein that plays a critical scaffolding role to link sarcomeres with organelles,12 that we have previously observed to be cleaved in Myh6-sTNF hearts,2 co-localizes with p62 and ubiquitin aggregates in cardiac myocytes. We considered that there were at least three possible mechanisms for the increased ubiquitin-protein conjugates in the Myh6-sTNF hearts: first, there may have been a decrease in proteasome mediated degradation of ubiquitin-protein conjugates; second, there could be impaired autophagy; third there could be an admixture of these mechanisms.
Proteasome Dysfunction in Myh6-sTNF Mouse Hearts
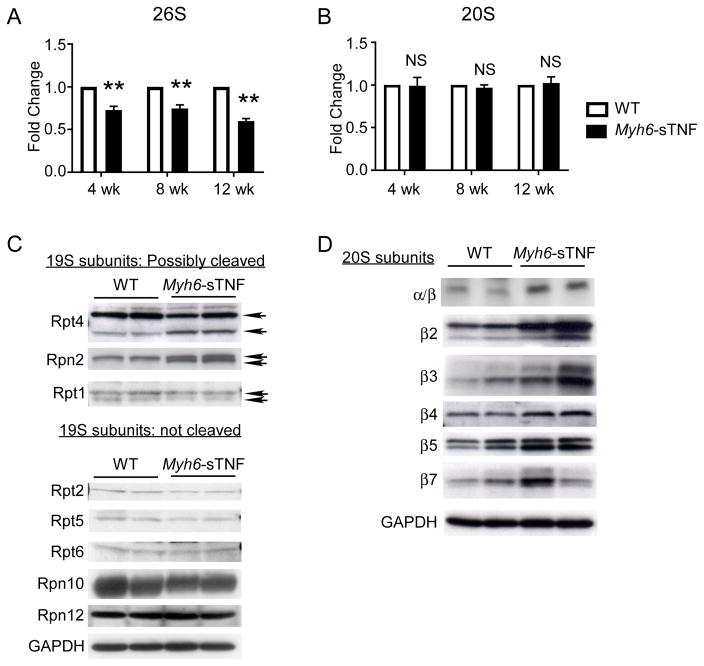
To determine whether alterations in proteasome function might have contributed to the increased ubiquitin-protein conjugates observed in the Myh6-sTNF hearts, we measured chymotrypsin-like activity of the myocardial 26S and the 20S proteasome fractions using an in vitro fluorescent assay. Figure 2A shows that 26S proteasome activity was decreased significantly in the Myh6-sTNF myocardial proteasome fractions when compared to the littermate controls from 4–12 weeks of age. However, function of the 20S core subunit was unimpaired through 12 weeks of age (Figure 2B), suggesting that dysfunction of the 19S cap was responsible for the observed decrease in activity of the 26S proteasome (Figure 2B). Interestingly when we measured expression levels of components of the 19S cap in Myh6-sTNF mouse hearts, we detected the presence of lower molecular weight species of the Rpt4 and Rpn2 subunits, suggesting potential cleavage of Rpt4 and Rpn2. We also detected the presence of a lower molecular weight species of Rpt1 in both the WT and Myh6-sTNF mice. Prior studies have shown that caspase-dependent cleavage of the 19S proteasome can lead to inhibition of the 26S proteasome.13, 14 Relevant to this discussion, we have previously reported increased activation of caspases 3,8, and 9 in the Myh6-sTNF mouse hearts.15, 16 Viewed together these data suggest (but do not prove) that the impaired function of the 19S cap observed in Myh6-sTNF mice may be related to cleavage of subunits of the 19S cap. To determine whether changes in chymotrypsin-like activity of the 26S subunit were accompanied by compensatory changes in the 20S proteasome subunit of the Myh6-sTNF mice, we also performed Western blot analyses of the 20S proteasome. Figure 2D shows increased expression of various components of the 20S proteasome suggesting that impairment of the 20S proteasome did not contribute to decreased activity of the 26S proteasome.
Figure 2.
Proteasome function in Myh6-sTNF hearts. A) The chymotrypsin-like activity of the 26S proteasome was assessed in 4, 8, and 12 weeks old Myh6-sTNF and littermate control mice by the cleavage of the fluorescent substrate Suc-LLVY-AMC in the presence of ATP. Results of the group data are shown as the ratio to the littermate control activity. B) The chymotrypsin-like activity of the 26S proteasome was assessed in 4, 8, and 12 weeks old Myh6-sTNF and littermate control mice (N=5) by the cleavage of the fluorescent substrate Suc-LLVY-AMC in the absence of ATP. Results of the group data shown as the ratio to the littermate control activity. **P<0.001, and NS = not significant. C) Representative Western blot analysis from 3 independent experiments of 19S protein subunits. Arrows indicate intact and cleaved subunits. D) Representative Western blot analysis of the core subunits of the 20S proteasome and individual catalytic subunits.
Impaired Autophagic Flux in Myh6-sTNF Mouse Hearts
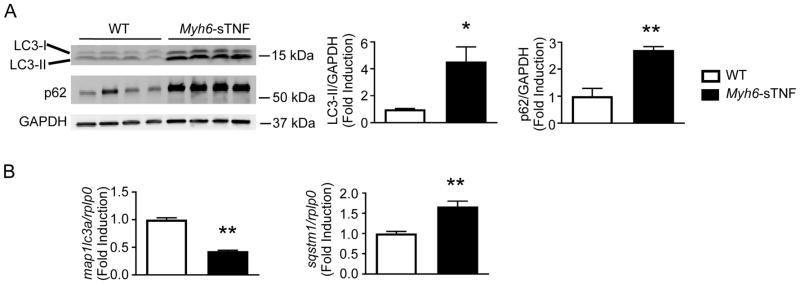
The autophagy-lysosome pathway acts as the back-up protein degradation pathway in the setting of proteasome dysfunction,17 and is primarily responsible for cleavage of a subset of long-lived proteins.18 The co-localization of p62 with ubiquitin in protein aggregates within cardiac myocytes (Figure 1C) suggested the possibility that autophagy was dysregulated in the Myh6-sTNF mouse hearts. To explore this possibility, we examined components of the autophagy-lysosome system. As shown in Figure 3A there were increased levels of LC3-I, the proteolytically processed form of LC3, and increased levels of LC3-II, the autophagosome-bound form of LC3. The accumulation of LC3-I and LC3-II did not appear to be secondary to increased transcription of LC3, insofar as the amount of mRNA (map1lc3a) was significantly decreased in the Myh6-sTNF mouse hearts (Figure 3B), suggesting that the observed changes in LC3-II protein levels were secondary to either increased formation (i.e., increased autophagy initiation), impaired clearance (i.e., impaired autophagic flux), or a combination of both. We also observed an increase in the amount of p62 protein in Myh6-sTNF mouse hearts when compared to littermate controls (Figure 3A), consistent with either increased transcription of p62 or impaired consumption of p62 during autophagy. Figure 3B shows that the mRNA levels for p62 (sqstm1) were increased in the 12 week Myh6-sTNF mouse hearts, indicating that increased transcription may have contributed to the increase in p62 protein levels.
Figure 3.
Accumulation of autophagy substrates in Myh6-sTNF hearts. A) Representative Western blot (left) and quantitative data (right) for LC3 and p62 levels in myocardial extracts from 12 weeks old WT and Myh6-sTNF mice. For quantitative analysis total of n=7 per group from two independent experiments. B) Quantitative RT-PCR for mRNA’s encoding LC3 (map1lc3a) and p62 (sqstm1). N=7 per group. *P<0.05 and **P<0.001.
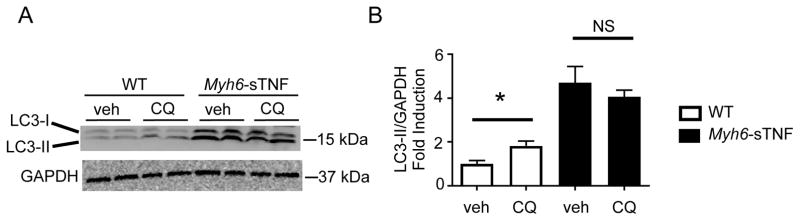
To determine whether the increased levels of LC3-II and p62 reflected impaired autophagic flux, we treated Myh6-sTNF mice with chloroquine, which inhibits lysosome acidification and fusion with autophagosomes, and then examined LC3-II accumulation in cardiac extracts as an indicator of autophagic flux.19 As shown in Figure 4A, short-term treatment with chloroquine (for 4 hours) resulted in an expected accumulation of LC3-II (a marker of autophagosome abundance) in LM control mice, reflecting intact autophagic flux. In contrast, there was no significant change in the levels of LC3-II in the chloroquine-treated Myh6-sTNF mice, suggesting that there was impaired clearance of autophagosomes. Viewed together with the increased levels of LC3-II and p62 in Myh6-sTNF mouse hearts this suggest that there is impaired autophagic flux.
Figure 4.
Impaired autophagic flux in Myh6-sTNF Mice. 12 weeks old WT littermates and Myh6-sTNF mice were treated with either vehicle (veh) or chloroquine (CQ) (40 mg/kg IP injection) 4 hours prior to sacrifice. A) Representative Western blot for LC3-II levels. B) Quantitative data including total of N=6 from multiple independent experiments. *P<0.05 and NS= not significant.
Mechanisms of Impaired Autophagic Flux in Myh6-sTNF Mouse Hearts
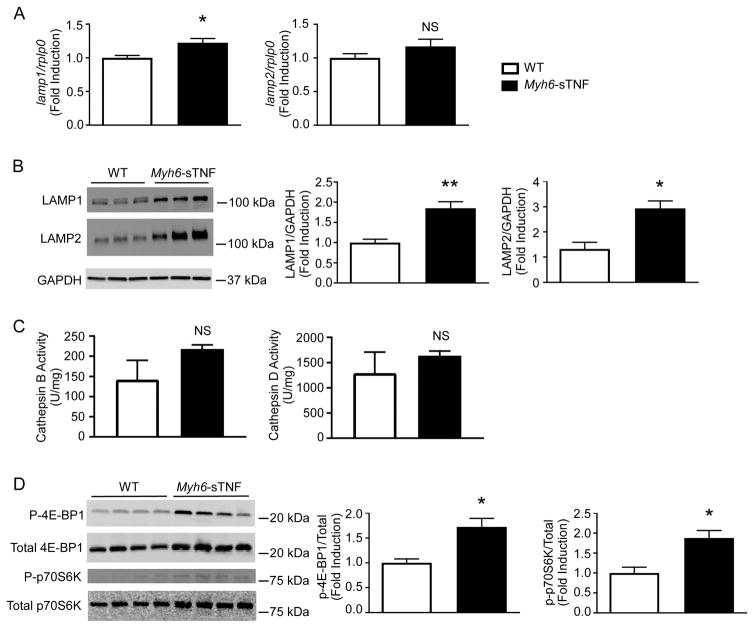
Several mechanisms have been linked to impaired autophagic flux in different cardiovascular disease states, including impaired lysosomal biogenesis,20 as well as lysosome dysfunction.21–24 Accordingly, to address whether lysosomal biogenesis was impaired, we measured levels of LAMP1 and LAMP2 mRNA and protein in the Myh6-sTNF mouse hearts. As shown in Figure 5A, there was a statistically significant increase in the level of lamp1 mRNA, and a non-significant increase in lamp2 mRNA levels. We also observed a significant increase in LAMP1 and LAMP2 protein levels in the Myh6-sTNF mouse hearts (Figure 5B). To characterize lysosome function we measured lysosomal cathepsin activity. There was no difference in cathepsin B or D activity in lysates from WT littermate and Myh6-sTNF mice (Figure 5C). Viewed together these data suggest that neither decreased lysosome abundance nor impaired lysosome function is responsible for the impaired autophagic flux in the Myh6-sTNF mouse hearts.
Figure 5.
Mechanism of impaired autophagy. All measurements were made in 12 weeks old WT and Myh6-sTNF mice. Quantitative analysis includes 7 mice per group. A) Lamp1 and lamp2 mRNA levels were measured by quantitative RT-PCR. B) Representative Western blot (left) and quantitative analysis (right) of LAMP1 and LAMP2 levels. C) Cathepsin B (left) and D (right) activity in WT and Myh6-sTNF mice. D) Representative Western blot (left) and quantitative analysis (right) measuring phosphorylated and total 4E-BP1 and p70S6K. Quantitative data are displayed as phosphorylated over total relative to WT. *P<0.05, **P<0.001, and NS= not significant.
Given the central role of mTOR in regulating autophagy,25 we next examined mTORC1 activity by measuring the phosphorylation of the downstream targets 4E-BP1 and p70S6K. As shown in Figure 5D there was increased phosphorylation of 4E-BP1 and p70S6K, suggesting that increased mTORC1 activity may have contributed to impaired autophagic flux in the Myh6-sTNF mouse hearts. In addition, we explored whether the observed impaired protein quality control in Myh6-sTNF hearts was accompanied by ER stress. As show in Supplemental Figure we observed increased protein levels for CHOP, ATF4, and ATF6 protein, as well as increased mRNA expression levels for ddit3 (CHOP) and sxbp1 (the spliced form for Xbp1), consistent with activation of the unfolded protein response (UPR).
Impaired Autophagy in Dilated Cardiomyopathy
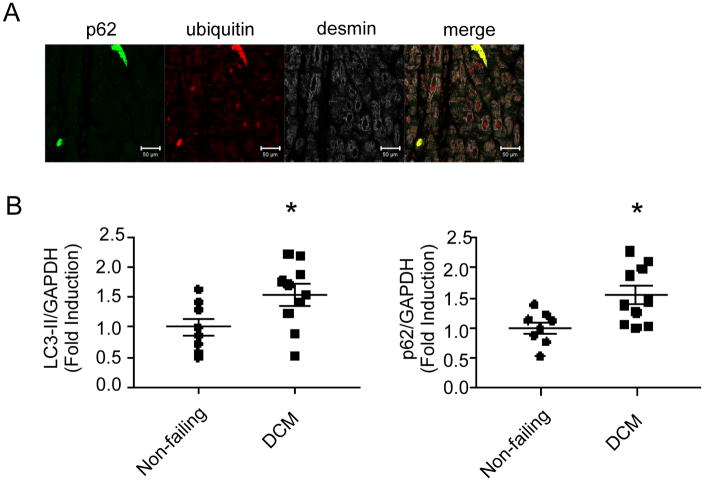
To determine whether the findings observed in the Myh6-sTNF mouse hearts were relevant to human heart failure, we performed immunofluorescence microscopy in hearts obtained from patients with dilated cardiomyopathy that were obtained at the time of cardiac transplantation. As shown in Figure 6A, we also observed the presence of aggregates containing both ubiquitin tagged proteins and p62 in the hearts of patients with dilated cardiomyopathy. We also observed that there were significantly increased levels of both LC3-II and p62 in dilated cardiomyopathy (Figure 6B).
Figure 6.
Accumulation of autophagy substrates in samples from patients with end-stage dilated cardiomyopathy (DCM). A) Representative immunofluorescence image from total of 9 individual DCM samples showing presence of protein aggregates containing ubiquitinated proteins and p62 (20× objective). B) Quantitative Western blot analysis of LC3 and p62 levels in samples from non-failing controls and patients with NICM. N=8 for non-failing and N=10 for DCM. *P<0.05.
DISCUSSION
The major finding of this study, in which we examined the mechanisms of protein quality control in a transgenic mouse model with cardiac restricted overexpression of TNF (Myh6-sTNF), is that sustained inflammatory signaling leads to the development of proteasome dysfunction and impaired autophagic flux. The following lines of evidence support these statements. First, we observed that there is a striking increase in ubiquitinated-protein conjugates in the hearts of the Myh6-sTNF mice (Figure 1), which was evident as early as 4 weeks of age, prior to the onset of cardiac remodeling.5 Moreover, the increase in ubiquitin-protein conjugates was maintained from 8 to 12 weeks age, at a time period when the Myh6-sTNF mouse hearts have been shown to undergo progressive LV dilation and LV wall thinning. Further, confocal immunofluorescence microscopy demonstrated that ubiquitinated proteins were incorporated into protein aggregates within individual cardiac myocytes in Myh6-sTNF mice, but were not detectable in WT mice. Second, we observed proteasome dysfunction, evidenced by decreased chymotrypsin-like activity of the 26S proteasome beginning at 4 weeks of age (Figure 2A and 2B). The decrease in activity of the 26S proteasome was secondary to dysfunction of the 19S proteasome, insofar as the chymotrypsin-like activity of the 20S proteasome was similar in the littermate controls and Myh6-sTNF mice. Our data suggest, but do not prove that the dysfunction of the 19S proteasome may be related to cleavage of 19S subunits (Figure 2C). Third, there was impaired autophagy in the Myh6-sTNF mouse hearts, leading to an accumulation of autophagy substrates, including increased levels of LC3-II and p62 (Figure 3A). Treatment with chloroquine indicated that impaired autophagic flux was responsible for the increased levels LC3-II and p62 (Figure 4). Although this study was not intended to delineate the mechanism(s) for impaired autophagy in the Myh6-sTNF mice, we did observe increased mTORC1 activation, a known inhibitor of autophagy, in the Myh6-sTNF mouse hearts (Figure 5D). Finally, to determine whether the findings observed in the Myh6-sTNF mouse hearts were relevant to human heart failure, we performed immunofluorescence microscopy in hearts obtained from patients with dilated cardiomyopathy that were obtained at the time of cardiac transplantation. As shown in Figure 6A, we also observed the presence of aggregates containing both ubiquitin tagged proteins and p62 in the hearts of patients with dilated cardiomyopathy, as well as increased levels of both LC3-II and p62 (Figure 6B), consistent with what we observed in the Myh6-sTNF mouse hearts.
Protein Quality Control in Dilated Cardiomyopathy
Protein homeostasis, or proteostasis, requires the coordinated action of multiple cellular networks. Under normal conditions these systems rapidly sense and rectify disturbances in the proteome in order to maintain tissue homeostasis. During stress these systems are transiently activated rapidly in order to preserve protein functionality and cell viability. However, in the setting of chronic stress, the cells homeostatic mechanisms are insufficient to maintain the proteostasis with the result that damaged proteins cannot be removed resulting in proteotoxicity and cell death. Proteotoxicity is a well-recognized aspect of neurodegenerative diseases. Increasingly it is also recognized to contribute to a variety of cardiovascular diseases.26, 27 In the setting of heart failure, proteotoxicity is best recognized as a component of desmin-related cardiomyopathy, and the relevant mechanisms have been studied in a mouse model of the disease. Desmin-related cardiomyopathy results from mutations in either desmin or the chaperone protein CryAB which result in protein misfolding leading to aggregate development, proteotoxicity, and cardiac dysfunction. In this setting in which the proteasome is dysfunctional, augmenting proteolytic activity decreases the accumulation of protein aggregates and delays disease progression.28–30 Autophagy plays a protective role in desmin related cardiomyopathy.31, 32 In tissue culture, the presence of aggregates provokes an increase in autophagy32, 33 which is a protective mechanism to remove cellular debris.26, 32, 34 However as the disease progresses autophagy becomes impaired, which then further contributes to impaired protein turnover and cardiac dysfunction.26, 31 Alternately, the accumulation of p62 bound to ubiquitinated proteins in cytosolic aggregates may prevent its physiologic targeting to the sarcomere, as has been reported previously.35 Indeed, although we observed that p62 was transcriptionally upregulated, it is possible that this upregulation is insufficient to permit a potential physiologic role for p62, because p62 remains sequestered within ubiqutinated protein aggregates in the myocyte.
Although the role of proteotoxicity in desmin-related cardiomyopathy has been well established, the effects of sustained inflammation on protein quality control are less well understood. Prior studies have demonstrated the accumulation of protein aggregates in two different transgenic mouse models of sustained activation of the TNF signaling pathway,1, 2 although the precise mechanisms for these observations have not been determined. Here we show that there is an accumulation of ubiquitin tagged proteins early in the disease which is due to both proteasome dysfunction and impaired autophagic flux. Although our results suggest impaired autophagic flux in setting of sustained TNF signaling, it should be noted that two prior studies have shown that acute stimulation with TNF stimulates autophagy in neonatal rat cardiac myocytes.8, 36 Thus, the effects of TNF on protein quality control are likely context dependent.
A number of mechanisms have been linked to impaired protein quality control in the heart. As noted, our data suggest that the decrease in activity of the 26S proteasome was secondary to a dysfunction of the 19S proteasome, insofar as the chymotrypsin-like activity of the 20S proteasome was similar in the littermate controls and Myh6-sTNF mice, which is consistent with the observations in desmin-related cardiomyopathy.28, 29 While activation of mTOR signaling has been demonstrated to inhibit proteasome function in some cell types, the mechanisms remain to be defined.18 Conceivably, cleavage of the proteasome subunits may be downstream of, or synergistic with the effects of mTOR activation in the Myh6-sTNF mice. In the present study, the accumulation of p62 and autophagosomes, as well as the decrease in autophagic flux observed with chloroquine treatment, suggests that there is impaired clearance of autophagosomes in Myh6-sTNF mice, pointing to impaired macro-autophagy as a mechanism for the increased accumulation of ubiquitinated proteins.17, 37 Although impaired lysosome biogenesis and function has previously been linked to decreased autophagy in the setting of ischemia reperfusion injury and doxorubicin cardiomyopathy,21, 24 this was not observed in the Myh6-sTNF mouse hearts insofar as neither lysosome abundance nor lysosome function were decreased in Myh6-sTNF mouse hearts. Our data do suggest that activation of mTORC1 may have been one of the mechanisms for the impaired autophagic flux observed in the Myh6-sTNF mice. While mTOR signaling has been ascribed a critical role in the homeostatic control of nutrient supply and stress-responses in the heart,38 the upstream pathways for mTOR activation in cardiomyopathy remain to be defined, and are the focus of active investigation. Viewed together, these results suggest the interesting possibility that inhibiting mTORC1 may improve protein quality control in inflammatory cardiomyopathies.
Conclusion
The results of this study show that sustained inflammatory signaling in the heart leads to the development of proteasome dysfunction and impaired autophagic flux, with the subsequent accumulation of ubiquitinated proteins. The loss of protein homeostasis in the heart may not only lead to proteotoxicity, but may also be important for a second reason. That is, it is possible that the loss of proteostasis may also contribute to impaired sarcomerogenesis, as well as maintenance of the highly ordered cytoskeletal structures that are required for normal contractility. Indeed, as heart failure advances there is a gradual loss of sarcomeres (myocytolysis),39 as well as loss of cytoskeletal elements.40 During reverse LV remodeling, there is an increase in sarcomerogenesis, as well as restoration of full length cytoskeletal elements. Interestingly, a recent study in conditional transgenic mouse model that develops reversible dilated cardiomyopathy showed that resolution of inflammation was accompanied by the clearance of protein aggregates in cardiac myocytes, improved alignment of sarcomeres, and improved contractile function.3 Germane to this discussion, genes involved in autophagy and the ubiquitin proteasome system were identified as two of the most heavily regulated groups during resolution of the inflammation-induced protein aggregates. Additional studies will be required to determine whether protein quality mechanisms are required for this response.
Supplementary Material
Acknowledgments
Sources of Funding: This research was supported by research funds from the NIH (RO1 HL58081, HL-73017-0, HL089543-01) to DLM and grants from NIH (HL107594) and Department of Veterans Affairs (1I01BX001969) to AD. JH received support from an NIH training grant (T32 HL007081) and the Oliver Langenberg Physician-Scientist Training Program.
Footnotes
Disclosures: None
References
- 1.Divakaran VG, Evans S, Topkara VK, Diwan A, Burchfield J, Gao F, Dong J, Tzeng HP, Sivasubramanian N, Barger PM, Mann DL. Tumor necrosis factor receptor-associated factor 2 signaling provokes adverse cardiac remodeling in the adult mammalian heart. Circ Heart Fail. 2013;6:535–43. doi: 10.1161/CIRCHEARTFAILURE.112.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panagopoulou P, Davos CH, Milner DJ, Varela E, Cameron J, Mann DL, Capetanaki Y. Desmin mediates TNF-alpha-induced aggregate formation and intercalated disk reorganization in heart failure. J Cell Biol. 2008;181:761–75. doi: 10.1083/jcb.200710049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topkara VK, Chambers KT, Yang KC, Tzeng HP, Evans S, Weinheimer C, Kovacs A, Robbins J, Barger P, Mann DL. Functional significance of the discordance between transcriptional profile and left ventricular structure/function during reverse remodeling. JCI Insight. 2016;1:e86038. doi: 10.1172/jci.insight.86038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat Rev Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 5.Sivasubramanian N, Coker ML, Kurrelmeyer KM, MacLellan WR, DeMayo FJ, Spinale FG, Mann DL. Left ventricular remodeling in transgenic mice with cardiac restricted overexpression of tumor necrosis factor. Circulation. 2001;104:826–31. doi: 10.1161/hc3401.093154. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Martinez C, Agell N, Llovera M, Lopez-Soriano FJ, Argiles JM. Tumour necrosis factor-alpha increases the ubiquitinization of rat skeletal muscle proteins. FEBS Lett. 1993;323:211–4. doi: 10.1016/0014-5793(93)81341-v. [DOI] [PubMed] [Google Scholar]
- 7.Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 8.Yang KC, Ma X, Liu H, Murphy J, Barger PM, Mann DL, Diwan A. Tumor necrosis factor receptor-associated factor 2 mediates mitochondrial autophagy. Circ Heart Fail. 2015;8:175–87. doi: 10.1161/CIRCHEARTFAILURE.114.001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, Diwan A, Lee JM. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 2014;34:9607–20. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–21. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su H, Wang X. p62 Stages an interplay between the ubiquitin-proteasome system and autophagy in the heart of defense against proteotoxic stress. Trends Cardiovasc Med. 2011;21:224–8. doi: 10.1016/j.tcm.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diokmetzidou A, Soumaka E, Kloukina I, Tsikitis M, Makridakis M, Varela A, Davos CH, Georgopoulos S, Anesti V, Vlahou A, Capetanaki Y. Desmin and alphaB-crystallin interplay in the maintenance of mitochondrial homeostasis and cardiomyocyte survival. J Cell Sci. 2016;129:3705–3720. doi: 10.1242/jcs.192203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adrain C, Creagh EM, Cullen SP, Martin SJ. Caspase-dependent inactivation of proteasome function during programmed cell death in Drosophila and man. J Biol Chem. 2004;279:36923–30. doi: 10.1074/jbc.M402638200. [DOI] [PubMed] [Google Scholar]
- 14.Sun XM, Butterworth M, MacFarlane M, Dubiel W, Ciechanover A, Cohen GM. Caspase activation inhibits proteasome function during apoptosis. Mol Cell. 2004;14:81–93. doi: 10.1016/s1097-2765(04)00156-x. [DOI] [PubMed] [Google Scholar]
- 15.Engel D, Peshock R, Armstong RC, Sivasubramanian N, Mann DL. Cardiac myocyte apoptosis provokes adverse cardiac remodeling in transgenic mice with targeted TNF overexpression. Am J Physiol Heart Circ Physiol. 2004;287:H1303–11. doi: 10.1152/ajpheart.00053.2004. [DOI] [PubMed] [Google Scholar]
- 16.Haudek SB, Taffet GE, Schneider MD, Mann DL. TNF provokes cardiomyocyte apoptosis and cardiac remodeling through activation of multiple cell death pathways. J Clin Invest. 2007;117:2692–701. doi: 10.1172/JCI29134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama S, Sou YS, Uemura T, Kametaka S, Saito T, Ishimura R, Kouno T, Bedford L, Mayer RJ, Lee MS, Yamamoto M, Waguri S, Tanaka K, Komatsu M. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J Biol Chem. 2014;289:24944–55. doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112:15790–7. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godar RJ, Ma X, Liu H, Murphy JT, Weinheimer CJ, Kovacs A, Crosby SD, Saftig P, Diwan A. Repetitive stimulation of autophagy-lysosome machinery by intermittent fasting preconditions the myocardium to ischemia-reperfusion injury. Autophagy. 2015;11:1537–60. doi: 10.1080/15548627.2015.1063768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, Weinheimer CJ, Barger PM, Diwan A. Regulation of the transcription factor EB-PGC1alpha axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol. 2015;35:956–76. doi: 10.1128/MCB.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–81. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim YC, Park HW, Sciarretta S, Mo JS, Jewell JL, Russell RC, Wu X, Sadoshima J, Guan KL. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun. 2014;5:4241. doi: 10.1038/ncomms5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan J, Mishra S, Qiu Y, Shi J, Trudeau K, Las G, Liesa M, Shirihai OS, Connors LH, Seldin DC, Falk RH, MacRae CA, Liao R. Lysosomal dysfunction and impaired autophagy underlie the pathogenesis of amyloidogenic light chain-mediated cardiotoxicity. EMBO Mol Med. 2014;6:1493–507. doi: 10.15252/emmm.201404190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li DL, Wang ZV, Ding G, Tan W, Luo X, Criollo A, Xie M, Jiang N, May H, Kyrychenko V, Schneider JW, Gillette TG, Hill JA. Doxorubicin Blocks Cardiomyocyte Autophagic Flux by Inhibiting Lysosome Acidification. Circulation. 2016;133:1668–87. doi: 10.1161/CIRCULATIONAHA.115.017443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLendon PM, Robbins J. Proteotoxicity and cardiac dysfunction. Circ Res. 2015;116:1863–82. doi: 10.1161/CIRCRESAHA.116.305372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis MS, Patterson C. Proteotoxicity and cardiac dysfunction--Alzheimer’s disease of the heart? N Engl J Med. 2013;368:455–64. doi: 10.1056/NEJMra1106180. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–26. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J Clin Invest. 2011;121:3689–700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, Hill JA, Sadoshima J, Robbins J. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–97. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–50. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–8. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 36.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–9. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–8. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 38.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–64. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hein S. Progression From Compensated Hypertrophy to Failure in the Pressure-Overloaded Human Heart: Structural Deterioration and Compensatory Mechanisms. Circulation. 2003;107:984–991. doi: 10.1161/01.cir.0000051865.66123.b7. [DOI] [PubMed] [Google Scholar]
- 40.Vatta M, Stetson SJ, Perez-Verdia A, Entman ML, Noon GP, Torre-Amione G, Bowles NE, Towbin JA. Molecular remodelling of dystrophin in patients with end-stage cardiomyopathies and reversal in patients on assistance-device therapy. The Lancet. 2002;359:936–941. doi: 10.1016/S0140-6736(02)08026-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.